2. 土壤与农业可持续发展国家重点实验室(中国科学院南京土壤研究所), 南京 210008;
3. 污染生态与环境工程重点实验室(中国科学院应用生态研究所), 沈阳 110164
2. State Key Laboratory of Soil and Sustainable Agriculture, Institute of Soil Science, Chinese Academy of Sciences, Nanjing 201008, China;
3. Key Laboratory of Pollution Ecology and Environmental Engineering, Institute of Applied Ecology, Chinese Academy of Sciences, Shenyang 110164, China
土壤具有高度的空间异质性,复杂的土壤孔隙结构为微生物提供了适合的生存空间,不同的微环境会影响微生物的丰富度[1],土壤理化性质沿土壤深度的变化会影响微生物的群落结构及互作。Bahram等[2]研究表明环境因素和微生物之间的竞争关系均会影响微生物群落的结构、组成和功能基因的丰度。深层土壤可能含有与表层土壤不同的且适应深层土壤环境的微生物群落[3-4],从而表现出不同的生态功能[5-6]。目前对于土壤微生物的研究大多数集中在0~20 cm的表层土壤上,对更深层土壤微生物群落空间变化的研究较少[7],对农业系统中土壤微生物群落随土壤深度变化的研究更少[8-9]。研究表明深层土壤微生物在土壤的形成、生物地球化学循环过程和污染物降解等方面发挥着重要的作用[10]。通过水分循环,深层土壤与表层土壤之间物质的流动和交换还会显著影响表层土壤质量从而影响表层植被的生产力[11]。土壤深度的增加会导致土壤微生物量和多样性的减少。Du等[12]发现由于土壤养分含量的不同导致微生物数量和香农指数均随深度而下降,且每层土壤中都含有特定的OTU(Operational taxonomic units)。Li等[13]通过使用磷脂脂肪酸分析方法(Phospholipid fatty acid,PLFA)研究稻田土壤中微生物群落随土壤深度的变化,表明土壤深度的增加,显著减少了土壤微生物的生物量和多样性,该结果意味着土壤含水量和养分含量是不同深度土壤微生物变化的主要驱动因子。土壤深度的增加还可能影响微生物的群落结构。Li等[14]的研究结果表明水稻土中细菌群落组成差异随深度的增加而变大,其中土壤全碳含量(Total carbon,TC)和全氮含量(Total nitrogen,TN)是导致水稻土细菌群落结构变化的主要影响因素。此外,土壤深度的增加还可能影响微生物的群落功能。Wang等[15]通过研究土壤氮循环相关基因与土壤深度之间的关系,表明除amoA-AOA基因外,其他氮循环相关功能基因丰度均随土壤深度的增加而显著降低,且这些基因与土壤碳、氮和碳氮比呈显著正相关。
过去对微生物的研究大多集中在物种数量和多样性上,而不是物种间的相互作用。然而在复杂的生态系统中,物种间的相互作用对生态系统的功能可能较物种丰富度和多样性更为重要[16-17]。微生物之间的相互作用是陆地生态系统中微生物群落互作网络的组成部分[18-19]。利用生态网络分析法(Ecological network analysis)可以揭示微生物潜在的互作机制及影响因素[20],是预测和改善土壤生态系统功能的重要步骤[21]。王丹丹等[22]使用生态网络分析法研究生物炭的添加对土壤根际真菌群落相互作用的影响。Purahong等[23]使用生态网络的分析方法研究了凋落物分解过程中细菌和真菌群落的交互作用模式,证明微生物群落变化和相互作用驱动了凋落物分解的不同阶段。
为揭示典型红壤水稻土剖面微生物分子生态网络特征,本研究选择江西鹰潭典型红壤水稻土作为实验对象,采集0~100 cm的5个剖面土壤,研究上层(0~20 cm),中层(20~60 cm)和下层(60~100 cm)不同深度红壤细菌和真菌的互作关系特征。通过对细菌16S rRNA和真菌18S rRNA的高通量测序,在前期研究的基础上[13-15],利用生态网络分析法构建三了个不同深度土壤微生物群落互作网络,以揭示红壤剖面不同深度微生物的界内及界间互作关系,并且利用随机森林和VPA两种分析方法探讨了影响微生物互作关系的环境因素。
1 材料与方法 1.1 供试材料供试土壤采集于江西鹰潭地区(东经116°54′~116°56′,北纬28°10′~28°13′,海拔34~62 m)晚稻收获后期(2014年10月至11月)。在该地区随机选择5个地点(每个地点相距至少3 km)采集5个重复样品,这些地点均具有土壤类型、耕作制度和气候条件等变量变化较小的特点。在每个地点,使用螺旋钻按照以下深度间隔(cm)垂直采集土壤样本:0~10、10~20、20~40、40~60、60~80和80~100,共30个样本[14]。
1.2 土壤理化性质的测定土壤含水率使用重量法测定。可溶性有机碳(Dissolved organic carbon,DOC)使用Multi N/C 3100分析仪(Analytitik Jena AG,Jena,Germany)测定。NH4+-N和NO3--N使用AA3 Continuous Flow Analyzer分析仪(Seal Analytic,Germany)测定。土壤全碳和全氮使用vario MICRO cube(Elementar Corporation,Germany)测定。
1.3 DNA提取及高通量测序使用FastDNA SPIK Kit土壤旋转试剂盒(MP Biomedicals,Solon,OH,USA)从0.5 g冻干土中提取土壤脱氧核糖核酸(DeoxyriboNucleic Acid,DNA)。通过1%的琼脂糖凝胶电泳和纳米滴分光光度计(NanoDrop,Wimmington,DE,USA),测定DNA的质量和浓度。测定合格的30个样品的DNA在-20℃下储存。
细菌16S rRNA的扩增引物序列是Bacteria-341F=5’-CCTACGGGAGGCAGCAG-3’,Bacteria-758R=5’-CTACCAGGGTATCTAATCC-3’。PCR扩增的条件为:变性95 ℃、10 s,退火55 ℃、30 s,延伸72 ℃、45 s,重复循环变性-退火-延伸45次。真菌18S rRNA的扩增引物序列是Fungal-FR1=5’-AICCATTCAATCGGTAIT-3’,Fungal-FR390=5’- CGATAACGAACGAGACCT-3’。PCR扩增的条件为:变性95 ℃、10 s,退火50 ℃、30 s,延伸72℃、45 s,重复循环变性-退火-延伸45次。
1.4 数据处理根据测序得到的细菌和真菌的OTU数据,使用Cytoscape软件中的CoNet插件构建微生物生态网络。分析步骤和网络参数的选择参考顾静馨[24]。网络分析中选择四种相关分析方法,即Pearson correlation,Spearman correlation,Bray-Curtis dissimilarity和Kullback-Leibler dissimilarity,起始连接数设置为500。然后,采用Benjamini-Hochberg方法标准化处理相关系数,即校正原有假设检验得到的显著性P值(P-value),并最终采用校正后的P值,保留P < 0.05的相关OTU构建关联网络。利用NetworkAnalyzer工具,获得网络的特征路径长度、连接数、节点数、群聚系数、网络密度和平均连通度等网络拓扑参数。
本研究通过R软件的‘vegan’数据包计算样品的丰富度和多样性。随机森林的分析方法是通过对数据集中的每一个解释变量分别随机置换以得到每个解释变量对被解释变量的贡献程度,以均方误差(mean square error,MSE)表示。本研究利用R软件中的‘randomForest’数据包进行随机森林模型的预测;对模型整体的检验利用‘rfUtilities’数据包;随机森林中每个变量对模型贡献程度的显著性检验利用‘rfPermute’数据包。方差分解分析(Variance Partitioning Analysis,VPA)是一种偏分析法,能够揭示不同环境因子对细菌真菌互作关系的相对贡献率大小,本研究使用R软件中的‘vegan’数据包完成。
2 结果 2.1 土壤理化性质沿深度的变化随着土壤深度的增加,土壤理化性质如pH、含水率(Soil moisture,SM)、TN、TC和碳氮比(C/N)等均显著变化(图 1)。例如,土壤pH先增加后减小,在40~60 cm之间土壤的pH最大;与之相反,NO3--N先减小后增加。此外,除DOC和NH4+-N-N的含量随深度的增加而增加外,TC、TN和C/N均随土壤深度的增加而减小。
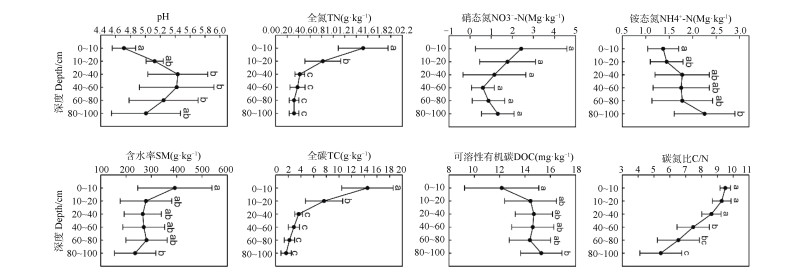
|
注:图中无相同小写字母表示理化性质在不同土壤深度中差异显著(P < 0.05)。下同Note:Different lowercase letters in the figure indicate significant difference of physicochemical properties among soil depth(P < 0.05). 图 1 不同深度土壤理化性质变化 Fig. 1 Physicochemical properties of the soil relative to soil depth |
沿土壤剖面计算了细菌和真菌群落的丰富度指数(图 2)。结果表明,尽管细菌和真菌的丰富度指数沿土壤深度的增加有相同的响应模式。但是相比于真菌,细菌丰富度对土壤深度的响应更加敏感。例如:细菌群落的丰富度指数随深度增加连续降低(P < 0.05);而真菌群落在60~100 cm深度范围内的丰富度并不随土壤深度的进一步增加而降低。该结果表明了相比于细菌而言,真菌群落对资源限制可能有更强的抵抗力。
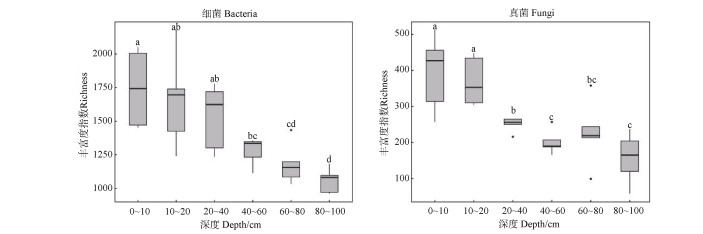
|
图 2 不同深度土壤细菌和真菌丰富度 Fig. 2 Richness of soil bacteria and fungi relative to soil depth |
基于红壤剖面细菌真菌丰富度指数的变化(图 2)及群落结构分布[14],本研究将0~100 cm土壤划分为上层(0~20 cm)、中层(20~60 cm)和下层(60~100 cm)。基于显著相关性对细菌和真菌的高通量测序数据构建互作网络(图 3),探究细菌-真菌群落的共现模式,使用NetworkAnalyzer工具计算网络的拓扑参数(表 1)。网络拓扑参数的分析表明,随着土壤深度的增加,网络的群聚系数、平均邻居数和网络密度均显著增加,特征路径长度和网络节点数则显著减小。该结果表明土壤微生物之间的总网络互作随深度的增加而变得复杂紧密。

|
a)上中下三层网络互相作用图Diagrams of interactions between the networks of the three soil layers. b)上中下三层网络微生物门水平上相互作用弦图Chord diagrams of the interactions between the networks of the three soil layers at the level of microbial phylum. 图 3 三个深度土壤微生物相互作用图 Fig. 3 Diagram of interactions between soil microbes relative to soil depth |
|
|
表 1 三个不同深度下微生物互作网络拓扑性质 Table 1 Topological properties of microbial interactions network relative to soil depth |
上中下三层土壤微生物互作网络图显示(图 3a),土壤微生物互作网络的模块化随深度增加,细菌、真菌群落界内的相互作用增加,其中细菌以正相互作用为主,真菌以负相互作用为主。上中下三层土壤微生物互作网络在门水平的弦图(图 3b)表明,细菌群落界内互作主要为酸杆菌门和绿弯菌门,真菌群落界内互作主要为子囊菌门和担子菌门。在上层网络中,细菌-真菌群落界间的互作复杂,包含多个细菌门类与真菌门类的互作;中层网络中,细菌-真菌群落界间的互作包含的门类减少,主要是酸杆菌门与子囊菌门之间的互作,绿弯菌门与子囊菌门之间的互作;下层网络中,细菌-真菌群落界间的相互作用进一步减少,细菌与真菌群落界内的相互作用增加,如酸杆菌门和子囊菌门的自相互作用增加。
进一步分析微生物界内和界间互作所占比例,细菌在互作网络中的占比随深度的增加而减少,真菌在互作网络中的占比增加;细菌、真菌群落界内互作与深度成正比。
不同深度红壤互作网络中群落之间连接数的分布情况显示(图 4),上层土壤的细菌、真菌群落界内互作和细菌-真菌群落界间互作分别占上层网络的37%、34%和29%,且均以正相互作用为主。中层土壤网络中以细菌群落界内正相互作用为主。下层土壤网络中以真菌群落界内负相互作用为主。研究结果进一步表明土壤中细菌-真菌群落界间互作与深度成反比,且细菌-真菌群落界间互作的正连接数占总连接数的占比随深度的增加显著减少(上层:18%,中层:6%,下层:3%)。

|
图 4 三个深度土壤细菌-真菌群落互作网络中相互作用关系 Fig. 4 Interactions in the soil bacterial and fungi community interaction network relative to soil depth |
利用随机森林进一步分析土壤微生物互作网络的拓扑性参数与土壤理化性质的关系(图 5),结果表明,细菌、真菌群落界内互作和细菌-真菌群落界间互作的主要影响因素是土壤中的TC含量(P < 0.05),其次为TN含量(P < 0.05)。且环境因子对细菌-真菌群落界间互作的解释量更大。
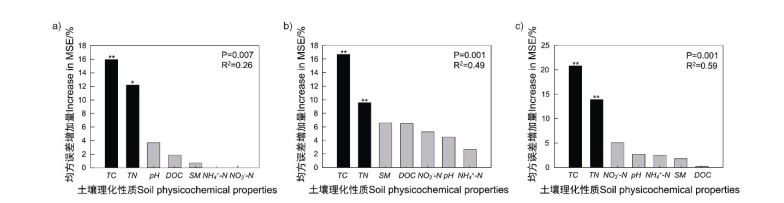
|
a)细菌群落界内互作Interactions within bacterial community. b)真菌群落界内互作Interactions within fungal community. c)细菌-真菌群落界间互作Interactions between bacteria and fungi communities. 图 5 土壤微生物互作网络与理化性质的随机森林 Fig. 5 Random forest analysis of physicochemical properties with soil microbial interaction network |
利用方差分解分析(Variance Partitioning Analysis,VPA)分析土壤pH、SM、C(TC和DOC)和N(TN、NO3--N和HN4+-N)对三层土壤微生物互作网络的贡献(图 6)。沿土壤剖面上中下三层微生物互作网络的总解释量分别为77.50%、92.29%和74.12%,土壤中的C和N是影响微生物互作网络的主要因素,且随着土壤深度的增加,土壤C的解释量从3.58%增至32.67%。VPA结果表明表层土壤微生物更加受到N的影响而深层土壤微生物更加受到土壤C的影响,随着深度的增加,影响微生物相互作用的环境因素由N转变为C。
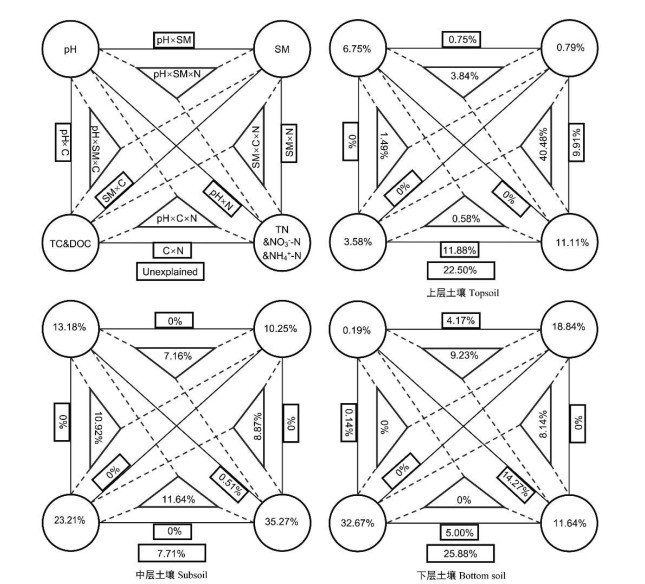
|
图 6 上中下三层土壤微生物互作网络VPA分析 Fig. 6 Variance partitioning analysis of soil microbial interaction network relative to soil depth |
本研究通过构建分子生态网络,探究了红壤水稻土剖面细菌、真菌群落界内及界间的相互作用及其影响因素。研究表明,尽管细菌和真菌的丰富度随土壤深度降低(图 2),但整体微生物互作网络的拓扑参数显著增强(表 1),表明相比于表层土壤,深层土壤微生物之间有更复杂的网络相互作用。这可能是由于农业系统中作物根系分泌物和地表凋落物主要增加了表层土壤中有机质的含量,并且施肥使表层土壤和深层土壤的养分差距变大,从而养分有效性随土壤深度不同而显著变化[25],导致表层土壤微生物多样性高而深层土壤微生物多样性低,微生物因土壤养分的减少而相互作用增加。这与Gu[26]和Li[27]等的研究结果一致—施肥会引起土壤性质的改变从而显著影响土壤微生物群落的组成及相互作用,深层土壤微生物互作网络较表层土壤微生物互作网络更加紧密复杂。资源的限制会导致细菌和真菌群落的相互作用由协作转变为竞争[19]。本研究中微生物之间的负相互作用随深度的增加而增加(图 3),这可能是由于土壤中碳和氮含量随深度增加而减少,从而导致微生物之间的竞争作用加强。Banerjee等[28]的研究结果表明,丰富的有机物和营养物会使微生物互作网络的负相互作用减少。
土壤微生物会借助趋化感应游向具有丰富营养物质的根际附近进行定殖与繁殖[29],丰富的资源环境有利于微生物的生长与物质交换。随着土壤深度的增加,土壤孔隙度减小,养分含量持续下降,影响了土壤微生物在充足营养源条件下的互作[30],导致细菌、真菌群落界内互作增加,而界间互作减少(图 4)。有研究指出,土壤高C/N有利于真菌生长,低C/N有利于细菌生长[29, 31-32]。本研究与之相反,在较高C/N的中层土壤主要以细菌群落界内的正相互作用为主,在较低C/N的深层土壤主要以真菌群落界内的负相互作用为主,可能是由于表层土壤中含有大量易降解有机碳而深层土壤中的有机碳以难降解有机碳为主[33-34],细菌更偏向于利用易降解有机碳,真菌更偏向于利用难降解有机碳[35-36],所以导致表层土壤更适合细菌生长而深层土壤更适合真菌生长。Huang等[37]的研究表明不同深度稻田土壤剖面,微生物群落利用的碳源种类不同。细菌-真菌群落界间互作不仅会受到环境因素的影响还会受到土壤中其它生物相互作用的影响[18]。本研究红壤细菌群落界内互作主要以正相互作用为主,真菌群落界内互作沿土壤剖面由正相互作用为主转变为负相互作用为主(图 4)。Eldridge等[38]的研究结果表明,土壤微生物网络中细菌和真菌的接连差异主要取决于土壤受到的干扰程度,环境干扰会增加微生物群落结构的不稳定性[39]。供试土壤是典型水稻土,施肥增加了表层土壤养分含量。微生物之间的相互作用主要是受到养分的驱动,丰富的资源会减少微生物之间的负相互作用[21, 28],随着土壤深度的增加,土壤养分含量减少,竞争性的相互作用在资源相对短缺时可能更加重要[21],所以真菌需要通过竞争性的相互作用来满足自身的需求。研究表明,根际土壤中的碳首先会被腐生真菌利用[40]。深层土壤中受到可利用碳组分的限制,细菌群落需要通过正相互作用来缓解环境的胁迫,同时扩大自己的生态位,以更好地获取资源[41]。
已有报道表明,微生物群落结构和功能随土壤剖面的变化显著受到土壤中TN和TOC的影响[42]。本研究对典型红壤水稻土的分析表明,土壤中C和N是影响细菌和真菌互作的主要因素(图 5),且随土壤深度的增加,对微生物互作网络的主要贡献因素由N转变为C(图 6)。Jangid等[43]的研究结果表明,微生物群落的差异与土壤可矿化的C、N含量和可提取的养分含量有关。Wan等[44]的研究结果表明,施氮肥会降低土壤C/N,进而改变土壤微生物群落结构。氮肥的施加会降低表层土壤微生物生物量及细菌/真菌的比例[45],所以N是表层土壤微生物互作的主要贡献因素。由于土壤C-N转化过程密切的偶联效应,土壤有机质分解受到土壤N养分的控制。氮肥的添加缓和了表层土壤中N对微生物的限制,减轻了微生物需要通过矿化惰性有机质以获取N的需求,使r策略微生物获得更强的竞争优势[46]。相比之下,深层土壤中可利用的碳源和氮源急剧减少,深层土壤微生物受到C的影响增加。Fierer等[7]的研究结果表明,土壤碳的可利用性随深度增加而下降,深层土壤微生物较表层土壤微生物更加受到土壤中C含量的限制,土壤中的C含量是细菌和真菌群落互作的主要驱动因素。
4 结论典型红壤水稻土微生物相互作用网络的拓扑性质如网络的连通度、群聚系数和网络密度等均与土壤的深度成正比;土壤深度越深,微生物的相互作用越紧密,模块化越明显,且细菌、真菌群落界内互作增加而细菌-真菌群落界间互作减弱。上层土壤以正相互作用为主,中层土壤以细菌群落界内正相互作用为主,下层土壤以真菌群落界内负相互作用为主。这主要受到土壤中C和N含量的影响,且随着土壤深度的增加,土壤中C对微生物互作网络影响显著增加,微生物互作网络最主要的贡献因素由N转变为C。
| [1] |
Bickel S, Or D. Soil bacterial diversity mediated by microscale aqueous-phase processes across biomes[J]. Nature Communications, 2020, 11: 116. DOI:10.1038/s41467-019-13966-w
(  0) 0) |
| [2] |
Bahram M, Hildebrand F, Forslund S K, et al. Structure and function of the global topsoil microbiome[J]. Nature, 2018, 560(7717): 233-237. DOI:10.1038/s41586-018-0386-6
(  0) 0) |
| [3] |
Bai R, Xi D, He J Z, et al. Activity, abundance and community structure of anammox bacteria along depth profiles in three different paddy soils[J]. Soil Biology & Biochemistry, 2015, 91: 212-221.
(  0) 0) |
| [4] |
Retzer J L, Lyon T L, Buckman H O, et al. The nature and properties of soils[J]. Journal of Range Management, 1952, 5(6): 420.
(  0) 0) |
| [5] |
Blume E, Bischoff M, Reichert J M, et al. Surface and subsurface microbial biomass, community structure and metabolic activity as a function of soil depth and season[J]. Applied Soil Ecology, 2002, 20(3): 171-181. DOI:10.1016/S0929-1393(02)00025-2
(  0) 0) |
| [6] |
Fritze H, Pietikäinen J, Pennanen T. Distribution of microbial biomass and phospholipid fatty acids in Podzol profiles under coniferous forest[J]. European Journal of Soil Science, 2000, 51(4): 565-573. DOI:10.1111/j.1365-2389.2000.00346.x
(  0) 0) |
| [7] |
Fierer N, Schimel J P, Holden P A. Variations in microbial community composition through two soil depth profiles[J]. Soil Biology & Biochemistry, 2003, 35(1): 167-176.
(  0) 0) |
| [8] |
Bao X L, Yu J, Liang W J, et al. The interactive effects of elevated ozone and wheat cultivars on soil microbial community composition and metabolic diversity[J]. Applied Soil Ecology, 2015, 87: 11-18. DOI:10.1016/j.apsoil.2014.11.003
(  0) 0) |
| [9] |
Yu C, Hu X M, Deng W, et al. Changes in soil microbial community structure and functional diversity in the rhizosphere surrounding mulberry subjected to long-term fertilization[J]. Applied Soil Ecology, 2015, 86: 30-40. DOI:10.1016/j.apsoil.2014.09.013
(  0) 0) |
| [10] |
Long P E, Williams K H, Hubbard S S, et al. Microbial metagenomics reveals climate-relevant subsurface biogeochemical processes[J]. Trends in Microbiology, 2016, 24(8): 600-610. DOI:10.1016/j.tim.2016.04.006
(  0) 0) |
| [11] |
Merrill S D, Liebig M A, Tanaka D L, et al. Comparison of soil quality and productivity at two sites differing in profile structure and topsoil properties[J]. Agriculture, Ecosystems & Environment, 2013, 179: 53-61.
(  0) 0) |
| [12] |
Du C, Geng Z C, Wang Q, et al. Variations in bacterial and fungal communities through soil depth profiles in a Betula albosinensis forest[J]. Journal of Microbiology, 2017, 55(9): 684-693. DOI:10.1007/s12275-017-6466-8
(  0) 0) |
| [13] |
Li X Y, Sun J, Wang H H, et al. Changes in the soil microbial phospholipid fatty acid profile with depth in three soil types of paddy fields in China[J]. Geoderma, 2017, 290: 69-74. DOI:10.1016/j.geoderma.2016.11.006
(  0) 0) |
| [14] |
Li X, Wang H H, Li X, et al. Shifts in bacterial community composition increase with depth in three soil types from paddy fields in China[J]. Pedobiologia, 2019, 77: 150589. DOI:10.1016/j.pedobi.2019.150589
(  0) 0) |
| [15] |
Wang H H, Li X, Li X, et al. Changes of microbial population and N-cycling function genes with depth in three Chinese paddy soils[J]. PLoS One, 2017, 12(12): e0189506. DOI:10.1371/journal.pone.0189506
(  0) 0) |
| [16] |
Zhou J Z, Deng Y, Luo F, et al. Functional molecular ecological networks[J]. mBio, 2010, 1(4): e00169-10. DOI:10.1128/mBio.00169-10
(  0) 0) |
| [17] |
Montoya J M, Pimm S L, Solé R V. Ecological networks and their fragility[J]. Nature, 2006, 442(7100): 259-264. DOI:10.1038/nature04927
(  0) 0) |
| [18] |
de Boer W. Upscaling of fungal-bacterial interactions: From the lab to the field[J]. Current Opinion in Microbiology, 2017, 37: 35-41. DOI:10.1016/j.mib.2017.03.007
(  0) 0) |
| [19] |
Deveau A, Bonito G, Uehling J, et al. Bacterial-fungal interactions: Ecology, mechanisms and challenges[J]. FEMS Microbiology Reviews, 2018, 42(3): 335-352. DOI:10.1093/femsre/fuy008
(  0) 0) |
| [20] |
Fuhrman J A. Microbial community structure and its functional implications[J]. Nature, 2009, 459(7244): 193-199. DOI:10.1038/nature08058
(  0) 0) |
| [21] |
de Menezes A B, Richardson A E, Thrall P H. Linking fungal-bacterial co-occurrences to soil ecosystem function[J]. Current Opinion in Microbiology, 2017, 37: 135-141. DOI:10.1016/j.mib.2017.06.006
(  0) 0) |
| [22] |
Wang D D, Yang Z P, Zhao Y, et al. Effect of biochar addition on the diversity and interaction of rhizosphere fungi in manure-fertilized soil (In Chinese)[J]. Environmental Science, 2018, 39(11): 5163-5169. [王丹丹, 杨泽平, 赵远, 等. 生物炭对施粪肥土壤中根际真菌群落多样性及相互作用的影响[J]. 环境科学, 2018, 39(11): 5163-5169.]
(  0) 0) |
| [23] |
Purahong W, Wubet T, Lentendu G, et al. Life in leaf litter: Novel insights into community dynamics of bacteria and fungi during litter decomposition[J]. Molecular Ecology, 2016, 25(16): 4059-4074. DOI:10.1111/mec.13739
(  0) 0) |
| [24] |
顾静馨. 土壤微生物生态网络的构建方法及其比较[D]. 江苏扬州: 扬州大学, 2015. Gu J X. The methods of constructing soil mircrobial ecological network and its comparison[D]. Yangzhou, Jiangsu: Yangzhou University, 2015. (  0) 0) |
| [25] |
Kaye J P, Hart S C. Competition for nitrogen between plants and soil microorganisms[J]. Trends in Ecology & Evolution, 1997, 12(4): 139-143.
(  0) 0) |
| [26] |
Gu Y F, Wang Y Y, Lu S G, et al. Long-term fertilization structures bacterial and archaeal communities along soil depth gradient in a paddy soil[J]. Frontiers in Microbiology, 2017, 8: 1516. DOI:10.3389/fmicb.2017.01516
(  0) 0) |
| [27] |
Li C H, Yan K, Tang L S, et al. Change in deep soil microbial communities due to long-term fertilization[J]. Soil Biology & Biochemistry, 2014, 75: 264-272.
(  0) 0) |
| [28] |
Banerjee S, Kirkby C A, Schmutter D, et al. Network analysis reveals functional redundancy and keystone taxa amongst bacterial and fungal communities during organic matter decomposition in an arable soil[J]. Soil Biology & Biochemistry, 2016, 97: 188-198.
(  0) 0) |
| [29] |
Wu L K, Lin X M, Lin W X. Advances and perspective in research on plant-soil-microbe interactions mediated by root exudates (In Chinese)[J]. Chinese Journal of Plant Ecology, 2014, 38(3): 298-310. [吴林坤, 林向民, 林文雄. 根系分泌物介导下植物-土壤-微生物互作关系研究进展与展望[J]. 植物生态学报, 2014, 38(3): 298-310.]
(  0) 0) |
| [30] |
Wu Z Y, Lin W X, Chen Z F, et al. Variations of soil microbial community diversity along an elevational gradient in mid-subtropical forest (In Chinese)[J]. Chinese Journal of Plant Ecology, 2013, 37(5): 397-406. [吴则焰, 林文雄, 陈志芳, 等. 中亚热带森林土壤微生物群落多样性随海拔梯度的变化[J]. 植物生态学报, 2013, 37(5): 397-406.]
(  0) 0) |
| [31] |
Bossuyt H, Denef K, Six J, et al. Influence of microbial populations and residue quality on aggregate stability[J]. Applied Soil Ecology, 2001, 16(3): 195-208. DOI:10.1016/S0929-1393(00)00116-5
(  0) 0) |
| [32] |
Six J, Frey S D, Thiet R K, et al. Bacterial and fungal contributions to carbon sequestration in agroecosystems[J]. Soil Science Society of America Journal, 2006, 70(2): 555-569. DOI:10.2136/sssaj2004.0347
(  0) 0) |
| [33] |
Franzluebbers A, Stuedemann J. Particulate and non-particulate fractions of soil organic carbon under pastures in the Southern Piedmont USA[J]. Environmental Pollution, 2002, 116: S53-S62. DOI:10.1016/S0269-7491(01)00247-0
(  0) 0) |
| [34] |
张燕. 黄土高原农田土壤碳和养分库分布及稳定性[D]. 陕西杨凌: 西北农林科技大学, 2017. Zhang Y. Composition and stability of soil carbon and nutrients reservoirs in farmland of the loess plateau[D]. Yangling, Shaanxi: Northwest A & F University, 2017. (  0) 0) |
| [35] |
Meidute S, Demoling F, Bååth E. Antagonistic and synergistic effects of fungal and bacterial growth in soil after adding different carbon and nitrogen sources[J]. Soil Biology & Biochemistry, 2008, 40(9): 2334-2343.
(  0) 0) |
| [36] |
丁建莉. 长期施肥对黑土微生物群落结构及其碳代谢的影响[D]. 北京: 中国农业科学院, 2017. Ding J L. Black soil microbial community structure and carbon metabolism in response to long-term fertilization[D]. Beijing: Chinese Academy of Agricultural Sciences, 2017. (  0) 0) |
| [37] |
Huang J, Sheng X F, He L Y, et al. Characterization of depth-related changes in bacterial community compositions and functions of a paddy soil profile[J]. FEMS Microbiology Letters, 2013, 347(1): 33-42. DOI:10.1111/1574-6968.12218
(  0) 0) |
| [38] |
Eldridge D J, Woodhouse J N, Curlevski N J A, et al. Soil-foraging animals alter the composition and co-occurrence of microbial communities in a desert shrubland[J]. The ISME Journal, 2015, 9(12): 2671-2681. DOI:10.1038/ismej.2015.70
(  0) 0) |
| [39] |
de Vries F T, Griffiths R I, Bailey M, et al. Soil bacterial networks are less stable under drought than fungal networks[J]. Nature Communications, 2018, 9: 3033. DOI:10.1038/s41467-018-05516-7
(  0) 0) |
| [40] |
Ballhausen M B, de Boer W. The sapro-rhizosphere: Carbon flow from saprotrophic fungi into fungus-feeding bacteria[J]. Soil Biology & Biochemistry, 2016, 102: 14-17.
(  0) 0) |
| [41] |
Freilich M A, Wieters E, Broitman B R, et al. Species co-occurrence networks: Can they reveal trophic and non-trophic interactions in ecological communities?[J]. Ecology, 2018, 99(3): 690-699. DOI:10.1002/ecy.2142
(  0) 0) |
| [42] |
于昊天. 鄱阳湖湿地剖面土壤微生物群落结构及功能的变化特征[D]. 南昌: 南昌大学, 2017. Yu H T. Characteristics of profile soil microbial community structure and function in Poyang lake wetland[D]. Nanchang: Nanchang University, 2017. (  0) 0) |
| [43] |
Jangid K, Williams M A, Franzluebbers A J, et al. Relative impacts of land-use, management intensity and fertilization upon soil microbial community structure in agricultural systems[J]. Soil Biology & Biochemistry, 2008, 40(11): 2843-2853.
(  0) 0) |
| [44] |
Wan X H, Huang Z Q, He Z M, et al. Soil C: N ratio is the major determinant of soil microbial community structure in subtropical coniferous and broadleaf forest plantations[J]. Plant and Soil, 2015, 387(1/2): 103-116.
(  0) 0) |
| [45] |
Rousk J, Brookes P C, Bååth E. Fungal and bacterial growth responses to N fertilization and pH in the 150-year 'Park Grass' UK grassland experiment[J]. FEMS Microbiology Ecology, 2011, 76(1): 89-99. DOI:10.1111/j.1574-6941.2010.01032.x
(  0) 0) |
| [46] |
Liang R B, Liang J, Qiao M F, et al. Effects of sim ulated exudate C: N stoichiometry on dynamics of carbon and microbial com-munity composition in a subalpine coniferous forest of western Sichuan, China (In Chinese)[J]. Chinese Journal of Plant Ecology, 2015, 39(5): 38-48. [梁儒彪, 梁进, 乔明锋, 等. 模拟根系分泌物C: N化学计量特征对川西亚高山森林土壤碳动态和微生物群落结构的影响[J]. 植物生态学报, 2015, 39(5): 38-48.]
(  0) 0) |
 2021, Vol. 58
2021, Vol. 58


