2. 土壤与农业可持续发展国家重点实验室(中国科学院南京土壤研究所), 南京 210008;
3. 浙江农林大学亚热带森林培育国家重点实验室, 杭州 311300;
4. 上海美吉生物医药科技有限公司, 上海 201318;
5. 南京农业大学生命科学学院, 南京 210095
2. State Key Laboratory of Soil and Sustainable Agriculture, Institute of Soil Science, Chinese Academy of Sciences, Nanjing 210008, China;
3. State Key Laboratory of Subtropical Silviculture, Zhejiang Agriculture and Forestry University, Hangzhou 311300, China;
4. Shanghai Majorbio Bio-pharm Biotechnology Co., Ltd, Shanghai 201318, China;
5. College of Life Sciences, Nanjing Agricultural University, Nanjing 210095, China
硝化作用被认为是全球氮循环的关键步骤,是由微生物驱动通过亚硝酸根,将氨氧化成硝酸根的过程[1]。而氮素作为森林生态系统中主要的限制性营养元素[2],其硝化作用产生的硝酸盐更易通过淋溶作用导致氮素流失和地下水污染等,此外硝酸根还可通过反硝化作用产生重要的温室气体氧化亚氮(N2O)。自1892年Winogradsky[3]发现氨氧化细菌(Ammonia-oxidizing bacteria,AOB)以来,人们一直认为硝化过程由两类微生物接力完成,分别为AOB催化的氨氧化过程和亚硝酸盐氧化细菌(Nitrite-oxidizing bacteria,NOB)催化的亚硝酸氧化过程,其中氨氧化过程被认为是硝化作用的限速步骤[4]。2005年,美国科学家David A. Stahl团队[5]从海洋中分离出第一株氨氧化古菌(Ammonia- oxidizing archaea,AOA),证明除细菌外,古菌也能够催化地球上的氨氧化过程。2015年,Daims[6]和van Kessel[7]等均发现了具有氨氧化活性的硝化螺旋细菌(Nitrospira),并证明其可将NH4+氧化成NO3-,称为完全硝化微生物(Complete ammonia oxidizer,comammox)。Comammox的发现打破了上百年来人们认为硝化过程必须由两类微生物接力完成的传统观念,使人们再次认识到硝化微生物的物种多样性和生理代谢机制的复杂。
酸性pH和低氨分子浓度是森林土壤的两个重要特征。一般认为酸性pH选择了严格嗜酸group 1.1a-associated AOA类群[8-10],且DNA-SIP(稳定性同位素示踪)实验证明该类AOA主导了森林土壤氨氧化过程[11]。但也有一些研究发现group 1.1b AOA可存在于酸性土壤[1, 12],且能主导某些酸性农田土壤氨氧化过程[13],以及group 1.1b AOA在酸性森林土壤中也广泛存在,且在转录组水平发挥了较高的活性[14-15]。尽管关于AOB适应酸性pH及其相对应的低NH3环境的机制目前多为推测[16-17],但不少森林土壤中确实存在β-proteobacterium(β-变形菌门)中Nitrosospira(亚硝化螺旋菌)属AOB,而Nitrosomonas(亚硝化单胞菌)AOB较少检测到[18-20]。最近日本科学家又从pH 3.0左右的酸性农田土壤中分离到耐酸γ-proteobacterium(γ-变形菌门)AOB菌株[21],然而森林土壤中是否存在耐酸γ-proteobacterium AOB仍不清楚。此外最近备受关注的comammox,其在森林土壤中生理生态的研究也刚刚起步,而已报道的comammox amoA基因PCR引物易产生非特异性扩增,可能导致其基于实时荧光定量PCR(qPCR)的丰度被严重高估[22]。土壤总DNA宏基因组测序由于不受PCR引物偏好性等限制,因此在土壤氨氧化微生物amoA基因相对丰度和群落组成的研究中更具优势。
马尾松林(Pinus massoniana)是我国南方广泛分布的主要人工林之一,具有高经济价值、耐干旱贫瘠等特点[23],其土壤为典型的酸性森林土壤。本文以浙江省建德市人工马尾松林土壤为研究对象,综合利用qPCR、凝胶电泳半定量和土壤总DNA宏基因组测序等手段,研究酸性森林土壤中AOA、AOB和comammox三种氨氧化微生物的丰度和群落组成等,以期加深对酸性森林生态系统中氨氧化微生物潜在生理生态学特征的认识。
1 材料与方法 1.1 供试材料土壤样品于2017年4月采自浙江省建德市人工马尾松林(119°28′E,29°48′N),该地区属亚热带海洋型季风气候,海拔约200 m,年均气温17.4 ℃,年均降水1 600 mm。土壤类型为凝灰岩发挥的红壤,选择3个林龄均为55年的马尾松林(间隔约2 000 m),分别标记为M1、M2和M3。五点法采集0~20 cm土壤,每个马尾松林采集3个样品,样品分成两份,一份保存于–20 ℃用于提取土壤总DNA,另一份4 ℃保存用于土壤理化性质测定。
1.2 土壤理化性质测定土壤含水量采用烘干法。pH采用电位法测量,水土比为5:1。有机碳含量通过浓硫酸-重铬酸钾法测定。全氮和碱解氮的含量分别采用半微量凯氏定氮法和碱解扩散法。有效磷测定采用0.05 mol·L–1 HCl和0.025 mol·L–1(1/2H2SO4)浸提—钼锑抗比色法。速效钾测定采用乙酸铵浸提—火焰光度法。经测定三个森林土壤pH为3.88~4.16,含水量173.1~203.4 g·kg–1,有机碳14.01~16.62 g·kg–1,全氮0.76~1.02 g·kg–1,碱解氮123.24~142.81 mg·kg–1,有效磷6.34~10.49 mg·kg–1,速效钾56.12~60.15 mg·kg–1。
1.3 土壤DNA提取土壤微生物总DNA提取采用Fast DNA Spin kit for soil试剂盒(MP Biomedicals,美国)。根据试剂盒操作说明提取土壤微生物总DNA,并使用Nanodrop ND-1000超微量分光光度计(NanoDrop Technologies,Wilmington,DE,美国)和1%的凝胶电泳检测DNA浓度和质量,将DNA于–20 ℃保藏。
1.4 实时荧光定量PCR和凝胶电泳半定量分析利用美国伯乐公司CFX96实时荧光定量PCR仪(Bio-Rad Laboratories,Hercules,美国)对人工马尾松林土壤中AOA、AOB和comammox的amoA基因进行定量分析。反应体系为20 μL:10 μL SYBR Premix Ex Taq(TaKaRa,日本),上下游引物0.5 μmol·L–1,2 μL(约1~10 ng)模板DNA,补充6 μL双蒸水。qPCR引物和反应条件见表 1[22, 24-26]。本实验每个样品设置三个重复,每个重复包含三个测试平行。将含有AOA、AOB和comammox amoA基因的单克隆质粒DNA梯度稀释成101~109的标准浓度作为qPCR标准曲线。通过琼脂糖凝胶电泳检查荧光定量PCR扩增产物的特异性。
|
|
表 1 定量PCR扩增引物及反应条件 Table 1 Quantitative PCR amplification primers and their reaction conditions |
本实验和已报道研究[27]均发现目前已知的comammox的amoA基因PCR引物易导致非特异性扩增,致使amoA基因丰度高估。因此利用凝胶电泳半定量法对qPCR结果进行数值初步矫正。即通过1.2%凝胶电泳和Bio-Rad公司的Quantity one v4.6.6软件检测待测样品qPCR产生的目标条带和非目标的相对灰度值,从而计算目标条带占泳道中总灰度值的百分比,进而用该百分比乘以qPCR检测结果,即得到样品中comammox的amoA基因实际拷贝数。同时我们将进一步用宏基因组中AOA、AOB和comammox三种amoA基因相对丰度,以及AOA和AOB的amoA基因的qPCR结果,进一步进行计算验证(见1.5)。
1.5 宏基因组测序和功能基因拼接及其相对丰度计算委托上海美吉生物对土壤总DNA进行150 bp双端测序(Illumina HiSeq 2 000)。每个样品分别产生约48Gb的原始数据,然后通过SeqPrep(https://github.com/jstjohn/SeqPrep)去除末端测序接头(adaptor),然后利用Sickle(https://github.com/najoshi/sickle)以默认参数对原始序列进行了质量控制和过滤,三个样品分别得到约42G bp高质量数据。使用metaSPAdes(version 3.10)宏基因组拼接软件对测序数据进行de novo从头组装,k-mer参数设置为21~29。使用Glimmer 3.0进行开放阅读框(ORFs)预测。以NCycDB数据库[28]中AOA和AOB的amoA基因以及Pjevac [22]、Daims [6]和Li[29]等文章中的comammox的amoA基因为参考序列,利用BLASTX与宏基因组拼接片段进行比对,提取E value ≤10–5且同源性高于70%的contigs作为amoA的候选序列,并进一步通过NCBI的nr数据库注释以及构建amoA进化树等手段进行验证去除错误注释的amoA基因序列。
分别采用两种方法计算宏基因组中AOA、AOB和comammox的amoA基因相对丰度。第一种方法,基于Mapped到amoA基因的HiSeq reads数统计。第二种方法,统计宏基因组中参与三种amoA contigs拼接所用的HiSeq reads数,reads数高低反映了该类基因的相对丰度高低。即根据amoA基因参考序列,通过BLASTX与原始reads进行比对,统计其中E value ≤10–5,同源性≥70%,blast hit区域大于25个氨基酸长度的reads数目,通过比较三种amoA基因mapping的reads总数,确定三者间的相对丰度。
1.6 系统发育进化树构建首先通过MOTHUR软件[30]按照95%序列同源性提取AOA、AOB和comammox的amoA基因的OTUs(operational taxonomic unit)代表性序列,然后利用MEGA 7.0软件的最大似然法(Maximum likelihood)构建三种氨氧化微生物amoA基因的进化树。
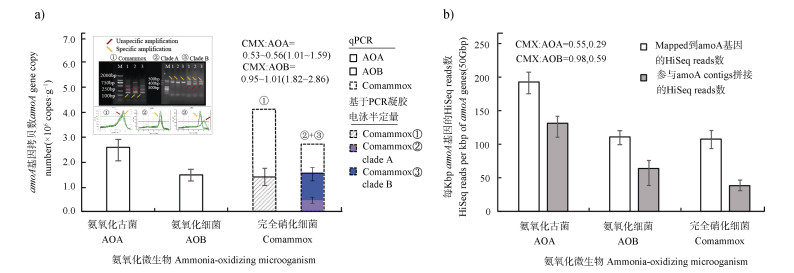
2 结果 2.1 AOA、AOB和comammox amoA基因丰度qPCR结果显示酸性马尾松林土壤中AOA和AOB的amoA基因丰度分别为2.61×106 copies·g–1和1.45×106 copies·g–1(图 1a)。基于引物Ntsp-amoA-162F/359R和引物comaA/B-244F/659R的qPCR结果显示comammox amoA基因拷贝数分别为4.14×106 copies·g–1和2.64×106 copies·g–1。然而,凝胶电泳图显示两组引物的qPCR结果均有明显的非特异性扩增,导致comammox的amoA基因qPCR结果的高估(图 1a)。通过凝胶电泳和Quantity one v4.6.6对comammox的amoA基因目标条带进行半定量估算后,得到两种引物扩增的comammox amoA基因总拷贝数分别为1.38×106 copies·g–1和1.47×106 copies·g–1(图 1a)。因此,氨氧化微生物功能基因amoA的相对丰度,comammox:AOA为0.53~0.56;comammox:AOB为0.95~1.01(图 1a)。
|
图 1 基于荧光定量PCR和凝胶电泳半定量(a)以及宏基因组数据(b)的AOA、AOB和comammox的amoA基因丰度 Fig. 1 amoA gene abundance of AOA, AOB and comammox based on qPCR, agarose gel electrophoresis semi-quantitative analysis (a) and metagenomic data (b) |
基于宏基因组Mapped到amoA基因的HiSeq reads数目和参与amoA contigs拼接的HiSeq reads数目分析发现,三类氨氧化微生物amoA基因相对丰度为:comammox:AOA约为0.55和0.29,comammox:AOB约为0.98和0.59(图 1b)。
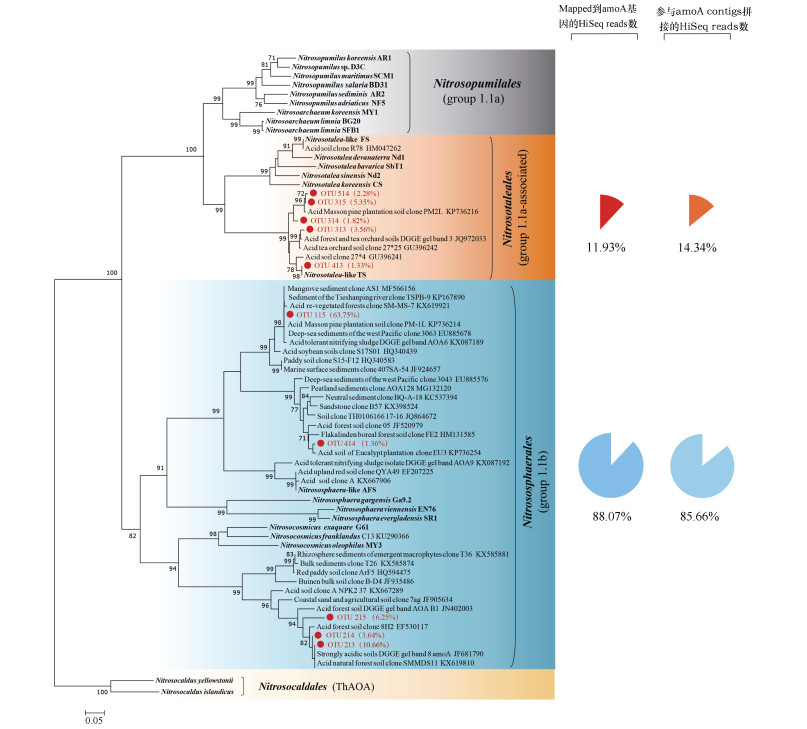
2.2 AOA、AOB和comammox的系统发育和群落组成目前AOA主要包括Nitrosopumilus(group 1.1a)、Nitrosotalea(group 1.1a-associated)、Nitrososphaera(group 1.1b)和Nitrosocaldus(ThAOA)[31]。宏基因组分析发现,马尾松林AOA其主要类群为group 1.1b,约占AOA总丰度的85.66%~88.07%,其次为group 1.1a-associated,占11.93%~14.34%,未检测到group 1.1a和Nitrosocaldus类群(图 2)。
|
图 2 基于宏基因组的氨氧化古菌(AOA)amoA基因系统发育树和相对丰度 Fig. 2 Phylogenetic tree and relative abundance of AOA amoA genes in the forest soil based on metagenomic analysis |
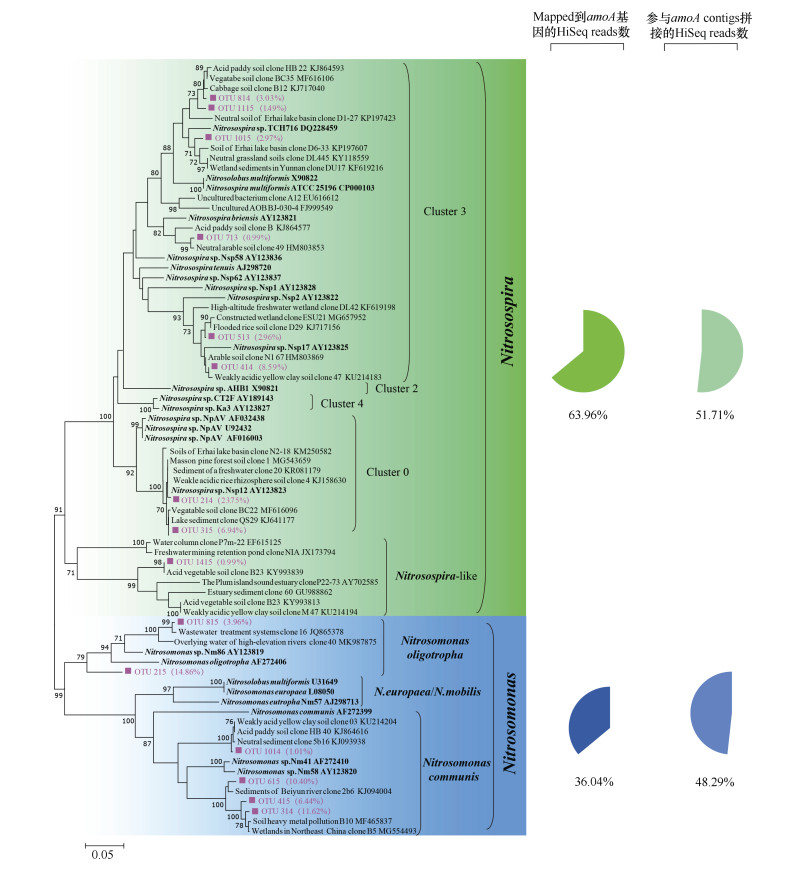
AOB主要分为β-AOB(Nitrosospira和Nitrosomonas)和γ-AOB(Nitrosococcus,亚硝化球菌属)[32]。其中Nitrosospira包含cluster 1~4、cluster 0、Nitrosospira sp. Nsp65和Nitrosospira sp. Nsp57/58等分支[33-34]。Nitrosomonas包含N. europaea/N. mobilis、N. communis、N. marina、N. oligotropha、N. cryotolerans、Nitrosomonas sp. Nm143和cluster 5等分支[35]。宏基因组分析显示,Nitrosospira是AOB的主要类群,占AOB amoA基因总丰度的51.71%~63.96%,主要隶属于cluster 3、0和Nitrosospira-like(图 3),而Nitrosomonas占36.04%~48.29%,主要隶属于Nitrosomonas communis和Nitrosomonas oligotropha(图 3)。
|
图 3 基于宏基因组的氨氧化细菌(AOB)amoA基因系统发育树和相对丰度 Fig. 3 Phylogenetic tree and relative abundance of AOB amoA genes in the forest soil based on metagenomic analysis |
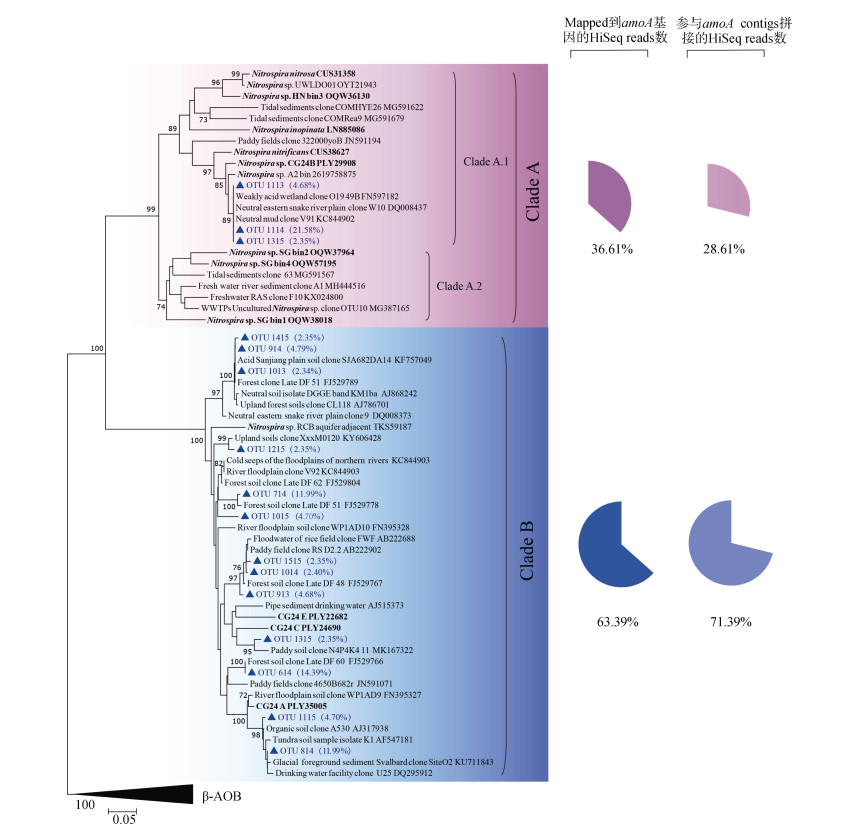
目前报道的comammox均来源于Nitrospira lineage Ⅱ,且主要分为clade A和clade B两支[6-7],近期有研究将comammox clade A进一步分为两个单系群,clade A.1和clade A.2[36]。土壤宏基因组分析发现,酸性森林土壤中comammox的主要类群为clade B,占comammox amoA基因总丰度的63.39%~71.39%,此外clade A.1占总序列数的28.61%~36.61%,未检测到clade A.2序列(图 4)。
|
图 4 基于宏基因组的完全硝化微生物(comammox)amoA基因系统发育树和相对丰度 Fig. 4 Phylogenetic tree and relative abundance of comammox amoA genes in the forest soil based on metagenomic analysis |
马尾松林土壤属于典型酸性森林土壤。为了得到AOA、AOB和comammox amoA基因准确丰度,本研究同时使用了qPCR、凝胶电泳半定量和宏基因组三种方法进行数据比较分析。qPCR和宏基因组均表明AOA amoA基因丰度约为AOB amoA基因的两倍(图 1)。而comammox amoA基因的两套引物存在明显的非特异扩增,该结果与Beach和Noguera[27]研究结果一致(图 1a)。通过凝胶电泳半定量对qPCR结果进行矫正以及宏基因组测序分析都发现,comammox amoA基因拷贝数和AOB的amoA基因处于同一个水平,均为AOA amoA基因拷贝数的50%左右(图 1)。然而,目前comammox的qPCR定量仍处于起步阶段。而凝胶电泳半定量也存在一定缺陷,其反映是qPCR结束时目标电泳条带和非目标电泳条带的灰度值之比,无法真正反映qPCR反应过程中特异扩增和非特异扩增释放的荧光信号强度之比。此外,凝胶电泳半定量目标条带位置,仍可能包含非特异扩增的序列。而宏基因组技术目前成本较高,数据分析也较繁琐。因此,仍需进一步发展comammox amoA基因qPCR更有效的引物,或选择comammox基因组中其他marker基因设计qPCR引物。
pH和NH3浓度是影响酸性土壤氨氧化微生物群落最重要的两个环境因子。自首株严格嗜酸AOA菌株分离以来[8],大量基于DNA-SIP实验证据表明group 1.1a-assocaited AOA主导了很多酸性土壤的氨氧化过程[9, 11, 37]。然而我们发现group 1.1b AOA也可以在某些酸性土壤中发挥着主导作用[13]。James I. Prosser团队[38-39]通过全球、区域和田块三个尺度研究发现group 1.1b AOA也存在于酸性土壤中,且亚枝cluster 11被认为嗜酸生长。本研究发现马尾松林土壤中group 1.1b AOA占据数量上的优势,而group 1.1a associated仅占12%(图 2)。此外,酸性土壤中NH3而被大量离子化成NH4+,而目前认为氨氧化微生物的底物为氨分子而非铵离子[40],因此推测酸性土壤中group 1.1b AOA能够适应较低氨环境。
酸性土壤中AOB的贡献长期以来一直存在较大争议。在AOA发现前,一直认为AOB驱动了酸性土壤的氨氧化过程。但后来发现分离于酸性土壤的Nitrosospira和Nitrosomonas纯菌株都不能在实验室内的酸性pH条件下生长[16]。Prosser等[16-17]推测可能有三种原因:(1)土壤中存在微域中性或碱性pH环境;(2)AOB在土壤中形成生物膜或团聚体抵抗酸性pH;(3)很多AOB菌株具有尿素水解酶活性,能够通过利用尿素抵抗(酸性土壤中)低氨胁迫。我们认为第三种解释可能混淆了酸性pH和低氨浓度两种环境胁迫,通过利用尿素确实可以应对低氨寡营养胁迫,但该AOB菌株能够在酸性pH条件下生存,还必须同时能够抵抗低pH胁迫。我们推测James I. Prosser等当时实验选择的Nitrosospira纯菌株本身能够耐受酸性pH,它只需要在此基础上通过利用尿素应对低氨胁迫即可在酸性pH培养基中生长。这点可以通过Prosser等[16-17]两篇文章中推测出来,五株来源于酸性土壤的AOB纯菌株均可以利用尿素,但只有一株Nitrosospira菌株可以通过利用尿素在酸性pH培养基中生长。因此,利用尿素对以上来源于酸性土壤的五株AOB纯菌株来说是适应酸性pH的必要但非充分条件。2017年Hayatsu等[21]分离得到在不添加尿素条件下,能够在酸性pH中生长的AOB纯菌株γ-proteobacterium TAO100。然而目前β-proteobacterium AOB适应酸性土壤的分子生物学机制仍不清楚。。
作为最新发现的comammox,其在地球各个生境中的分布规律备受关注。目前发现comammox在土壤、沉积物、滩涂、植物叶表、地下水、饮用水以及废水处理系统均有分布[6-7, 22, 36]。本研究发现酸性森林土壤也存在丰度与AOB(每个AOB细胞平均含有1.5个拷贝的amoA基因)相当的comammox类群(图 1),其中clade B占comammox总类群的64%,而clade A约占36%。该结果和已报到森林土壤中comammox群落组成基本一致[22, 36]。不同的是,Xia等[36]发现森林土壤中clade A主要隶属于clade A.2分支,而本研究发现马尾松林土壤中clade A主要隶属于clade A.1(图 4)。以上数据暗示comammox无论在clade B还是clade A类群中都存在能够适应酸性土壤环境的物种。Michael Wagner课题组[41]继2015年发现第一株comammox纯菌株后,也明确了已知的comammox纯菌株对底物氨具有很高的亲和力,其亲和力高于所有已知AOB菌株和绝大部分土壤来源的AOA菌株,因此这可以从一定程度解释comammox如何适应酸性土壤中的低NH3胁迫。然而,comammox如何适应酸性pH胁迫仍不清楚。
4 结论本文利用qPCR、凝胶电泳半定量和宏基因组测序等技术研究酸性森林土壤氨氧化微生物的群落结构。qPCR显示AOA和AOB的amoA基因丰度为2.61×106 copies·g–1和1.45×106 copies·g–1。而comammox amoA基因的扩增引物产生明显的非特异性扩增,导致qPCR结果的高估。半定量和宏基因组分析发现,氨氧化微生物amoA基因相对丰度为:comammox:AOA=0.55,comammox:AOB=0.98,推算comammox amoA基因真实丰度为(1.38~1.47)×106 copies·g–1。此外,AOA主要类群为group 1.1b占其总丰度的88.07%,而经典嗜酸类群group 1.1a-associated仅占11.93%。AOB主要类群为Nitrosospira(63.96%),而Nitrosomonas为36.04%。comammox主要类群为clade B(63.39%),而clade A仅占36.61%且均隶属于clade A.1亚枝。
致谢 感谢浙江农林大学孙凯和张云晴等同学在样品采集和理化性质测试中给予的帮助!
| [1] |
He J Z, Shen J P, Zhang L M, et al. Quantitative analyses of the abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea of a Chinese upland red soil under long-term fertilization practices[J]. Environmental Microbiology, 2007, 9(9): 2364-2374. DOI:10.1111/j.1462-2920.2007.01358.x
(  0) 0) |
| [2] |
Rennenberg H, Dannenmann M, Gessler A, et al. Nitrogen balance in forest soils: Nutritional limitation of plants under climate change stresses[J]. Plant Biology, 2009, 11: 4-23. DOI:10.1111/j.1438-8677.2009.00241.x
(  0) 0) |
| [3] |
Winogradsky S. Contributions a la morphologie des organisms de la nitrification[J]. Arkhiv Biologischeski Nauk(St. Petersbourg), 1892, 1: 87-137.
(  0) 0) |
| [4] |
Gubry-Rangin C, Nicol G W, Prosser J I. Archaea rather than bacteria control nitrification in two agricultural acidic soils[J]. FEMS Microbiology Ecology, 2010, 74(3): 566-574. DOI:10.1111/j.1574-6941.2010.00971.x
(  0) 0) |
| [5] |
Könneke M, Bernhard A E, de la Torre J R, et al. Isolation of an autotrophic ammonia-oxidizing marine archaeon[J]. Nature, 2005, 437(7058): 543-546. DOI:10.1038/nature03911
(  0) 0) |
| [6] |
Daims H, Lebedeva E V, Pjevac P, et al. Complete nitrification by Nitrospira bacteria[J]. Nature, 2015, 528(7583): 504-509. DOI:10.1038/nature16461
(  0) 0) |
| [7] |
van Kessel M A H J, Speth D R, Albertsen M, et al. Complete nitrification by a single microorganism[J]. Nature, 2015, 528(7583): 555-559. DOI:10.1038/nature16459
(  0) 0) |
| [8] |
Lehtovirta-Morley L E, Stoecker K, Vilcinskas A, et al. Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil[J]. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(38): 15892-15897. DOI:10.1073/pnas.1107196108
(  0) 0) |
| [9] |
Zhang L M, Hu H W, Shen J P, et al. Ammonia-oxidizing archaea have more important role than ammonia-oxidizing bacteria in ammonia oxidation of strongly acidic soils[J]. The ISME Journal, 2012, 6(5): 1032-1045. DOI:10.1038/ismej.2011.168
(  0) 0) |
| [10] |
He J Z, Hu H W, Zhang L M. Current insights into the autotrophic thaumarchaeal ammonia oxidation in acidic soils[J]. Soil Biology & Biochemistry, 2012, 55: 146-154.
(  0) 0) |
| [11] |
Lu, Han W Y, Zhang J B, et al. Nitrification of archaeal ammonia oxidizers in acid soils is supported by hydrolysis of urea[J]. The ISME Journal, 2012, 6(10): 1978-1984. DOI:10.1038/ismej.2012.45
(  0) 0) |
| [12] |
Wang B Z, Qin W, Ren Y, et al. Expansion of Thaumarchaeota habitat range is correlated with horizontal transfer of ATPase operons[J]. The ISME Journal, 2019, 13(12): 3067-3079. DOI:10.1038/s41396-019-0493-x
(  0) 0) |
| [13] |
Wang B Z, Zheng Y, Huang R, et al. Active ammonia oxidizers in an acidic soil are phylogenetically closely related to neutrophilic archaeon[J]. Applied and Environmental Microbiology, 2014, 80(5): 1684-1691. DOI:10.1128/AEM.03633-13
(  0) 0) |
| [14] |
Stempfhuber B, Engel M, Fischer D, et al. pH as a driver for ammonia-oxidizing archaea in forest soils[J]. Microbial Ecology, 2015, 69(4): 879-883. DOI:10.1007/s00248-014-0548-5
(  0) 0) |
| [15] |
Wu R N, Meng H, Wang Y F, et al. A more comprehensive community of ammonia-oxidizing archaea(AOA)revealed by genomic DNA and RNA analyses of amoA gene in subtropical acidic forest soils[J]. Microbial Ecology, 2017, 74(4): 910-922. DOI:10.1007/s00248-017-1045-4
(  0) 0) |
| [16] |
Allison S M, Prosser J I. Urease activity in neutrophilic autotrophic ammonia-oxidizing bacteria isolated from acid soils[J]. Soil Biology & Biochemistry, 1991, 23(1): 45-51.
(  0) 0) |
| [17] |
Burton S A Q, Prosser J I. Autotrophic ammonia oxidation at low pH through urea hydrolysis[J]. Applied and Environmental Microbiology, 2001, 67(7): 2952-2957. DOI:10.1128/AEM.67.7.2952-2957.2001
(  0) 0) |
| [18] |
Mintie A T, Heichen R S, Cromack K Jr, et al. Ammonia-oxidizing bacteria along meadow-to-forest transects in the Oregon Cascade Mountains[J]. Applied and Environmental Microbiology, 2003, 69(6): 3129-3136. DOI:10.1128/AEM.69.6.3129-3136.2003
(  0) 0) |
| [19] |
Long X E, Chen C R, Xu Z H, et al. Abundance and community structure of ammonia oxidizing bacteria and Archaea in a Sweden boreal forest soil under 19-year fertilization and 12-year warming[J]. Journal of Soils and Sediments, 2012, 12(7): 1124-1133. DOI:10.1007/s11368-012-0532-y
(  0) 0) |
| [20] |
Hastings R C, Butler C, Singleton I, et al. Analysis of ammonia-oxidizing bacteria populations in acid forest soil during conditions of moisture limitation[J]. Letters in Applied Microbiology, 2000, 30(1): 14-18. DOI:10.1046/j.1472-765x.2000.00630.x
(  0) 0) |
| [21] |
Hayatsu M, Tago K, Uchiyama I, et al. An acid-tolerant ammonia-oxidizing γ-proteobacterium from soil[J]. The ISME Journal, 2017, 11(5): 1130-1141. DOI:10.1038/ismej.2016.191
(  0) 0) |
| [22] |
Pjevac P, Schauberger C, Poghosyan L, et al. AmoA-targeted polymerase chain reaction primers for the specific detection and quantification of comammox Nitrospira in the environment[J]. Frontiers in Microbiology, 2017, 8: 1508. DOI:10.1101/096891
(  0) 0) |
| [23] |
Zhang K R, Liu Y D, Zhu X W, et al. Community types and species diversity of Pinus massoniana forests of Yuelu Mountain, Changsha (In Chinese)[J]. Scientia Silvae Sinicae, 2011, 47(4): 86-94. [张克荣, 刘应迪, 朱晓文, 等. 长沙岳麓山马尾松林的群落类型划分及物种多样性分析[J]. 林业科学, 2011, 47(4): 86-94.]
(  0) 0) |
| [24] |
Francis C A, Roberts K J, Beman J M, et al. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean[J]. Proceedings of the National Academy of Sciences of the United States of America, 2005, 102(41): 14683-14688. DOI:10.1073/pnas.0506625102
(  0) 0) |
| [25] |
Rotthauwe J H, Witzel K P, Liesack W. The ammonia monooxygenase structural gene amoA as a functional marker: Molecular fine-scale analysis of natural ammonia-oxidizing populations[J]. Applied and Environmental Microbiology, 1997, 63(12): 4704-4712. DOI:10.1128/aem.63.12.4704-4712.1997
(  0) 0) |
| [26] |
Fowler S J, Palomo A, Dechesne A, et al. Comammox Nitrospira are abundant ammonia oxidizers in diverse groundwater-fed rapid sand filter communities[J]. Environmental Microbiology, 2018, 20(3): 1002-1015. DOI:10.1111/1462-2920.14033
(  0) 0) |
| [27] |
Beach N K, Noguera D R. Design and assessment of species-level qPCR primers targeting comammox[J]. Frontiers in Microbiology, 2019, 10: 36. DOI:10.3389/fmicb.2019.00036
(  0) 0) |
| [28] |
Tu Q C, Lin L, Cheng L, et al. NCycDB: A curated integrative database for fast and accurate metagenomic profiling of nitrogen cycling genes[J]. Bioinformatics, 2019, 35(6): 1040-1048. DOI:10.1093/bioinformatics/bty741
(  0) 0) |
| [29] |
Li C Y, Hu H W, Chen Q L, et al. Comammox Nitrospira play an active role in nitrification of agricultural soils amended with nitrogen fertilizers[J]. Soil Biology & Biochemistry, 2019, 138: 107609.
(  0) 0) |
| [30] |
Schloss P D, Westcott S L, Ryabin T, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities[J]. Applied and Environmental Microbiology, 2009, 75(23): 7537-7541. DOI:10.1128/AEM.01541-09
(  0) 0) |
| [31] |
Pester M, Rattei T, Flechl S, et al. AmoA-based consensus phylogeny of ammonia-oxidizing archaea and deep sequencing of amoA genes from soils of four different geographic regions[J]. Environmental Microbiology, 2012, 14(2): 525-539. DOI:10.1111/j.1462-2920.2011.02666.x
(  0) 0) |
| [32] |
Head I M, Hiorns W D, Embley T M, et al. The phylogeny of autotrophic ammonia-oxidizing bacteria as determined by analysis of 16S ribosomal RNA gene sequences[J]. Journal of General Microbiology, 1993, 139(6): 1147-1153. DOI:10.1099/00221287-139-6-1147
(  0) 0) |
| [33] |
Stephen J R, McCaig A E, Smith Z, et al. Molecular diversity of soil and marine 16S rRNA gene sequences related to beta-subgroup ammonia-oxidizing bacteria[J]. Applied and Environmental Microbiology, 1996, 62(11): 4147-4154. DOI:10.1128/aem.62.11.4147-4154.1996
(  0) 0) |
| [34] |
Kowalchuk G A, Stephen J R, de Boer W, et al. Analysis of ammonia-oxidizing bacteria of the beta subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments[J]. Applied and Environmental Microbiology, 1997, 63(4): 1489-1497. DOI:10.1128/aem.63.4.1489-1497.1997
(  0) 0) |
| [35] |
Prosser J I, Head I M, Stein L Y. The family Nitrosomonadaceae[M]//The prokaryotes. Berlin, Heidelberg: Springer Berlin Heidelberg, 2014: 901-918.
(  0) 0) |
| [36] |
Xia F, Wang J G, Zhu T, et al. Ubiquity and diversity of complete ammonia oxidizers(comammox)[J]. Applied and Environmental Microbiology, 2018, 84(24): e01390-18. DOI:10.1128/aem.01390-18
(  0) 0) |
| [37] |
Lu, Jia Z J. Urease gene-containing Archaea dominate autotrophic ammonia oxidation in two acid soils[J]. Environmental Microbiology, 2013, 15(6): 1795-1809. DOI:10.1111/1462-2920.12071
(  0) 0) |
| [38] |
Gubry-Rangin C, Hai B, Quince C, et al. Niche specialization of terrestrial archaeal ammonia oxidizers[J]. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(52): 21206-21211. DOI:10.1073/pnas.1109000108
(  0) 0) |
| [39] |
Gubry-Rangin C, Kratsch C, Williams T A, et al. Coupling of diversification and pH adaptation during the evolution of terrestrial Thaumarchaeota[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(30): 9370-9375. DOI:10.1073/pnas.1419329112
(  0) 0) |
| [40] |
Stahl D A, de la Torre J R. Physiology and diversity of ammonia-oxidizing Archaea[J]. Annual Review of Microbiology, 2012, 66(1): 83-101. DOI:10.1146/annurev-micro-092611-150128
(  0) 0) |
| [41] |
Kits K D, Sedlacek C J, Lebedeva E V, et al. Kinetic analysis of a complete nitrifier reveals an oligotrophic lifestyle[J]. Nature, 2017, 549(7671): 269-272. DOI:10.1038/nature23679
(  0) 0) |
 2021, Vol. 58
2021, Vol. 58


