2. 中国科学院大学, 北京 100049;
3. 农业农村部黄淮海平原农业环境重点实验室/山东省环保肥料工程技术研究中心, 山东省农业科学院农业资源与环境研究所, 济南 250100;
4. 国家红壤改良工程技术研究中心, 江西省红壤研究所, 南昌 330046
2. University of Chinese Academy of Sciences, Beijing 100049, China;
3. Key Laboratory of Agro-Environment of Huang-Huai-Hai Plain, Ministry of Agriculture and Rural Affairs/Shandong Environmental Fertilizer Engineering Technology Research Center, Institute of Agricultural Resources and Environment, Shandong Academy of Agricultural Sciences, Jinan 250100, China;
4. Jiangxi Institute of Red Soil, National Engineering and Technology Research Center for Red Soil Improvement, Nanchang 330046, China
±壤反硝化是±壤氮循环的重要过程,是温室气体氧化亚氮(N2O)的重要排放源[1]。±壤潜在反硝化作用的大小可用±壤反硝化潜势(Soil denitrification potential,SDP)来表示,常用的测定方法中,乙炔抑制厌氧培养法由于经济且易操作性应用最广泛。±壤反硝化包括生物反硝化和化学反硝化,其中以生物反硝化为主[2]。生物反硝化是指异养微生物在厌氧条件下将NO3–逐步还原为N2的硝酸盐异化过程[3]。反硝化细菌群落是一个复杂的生物体系,广泛分布于复杂的属种中,而不是一个特定的分类群。因此16rRNA并不适用于反硝化微生物的系统发生研究,而功能基因经常被用来表征反硝化微生物,如narG、nirS、nirK和nosZ基因经常被广泛用于反硝化细菌群落的丰度和结构的检测[4-5]。传统的微生物鉴定技术并不适用于反硝化微生物的鉴定[6]。高通量测序技术一次可以对几十万到几百万条DNA分子进行测序[7],与其他分子生物鉴定技术相比具有通量高、节省成本和时间的优点,因此,越来越多的被应用于反硝化微生物的鉴定。由NO2–到NO的反应被认为是区分反硝化过程与其他硝酸盐代谢的关键步骤,也被认为是反硝化过程的限速步骤[8-9]。nirS/nirK基因是控制此过程的编码基因[10],但nirS型微生物较nirK型微生物在环境中分布更广泛,因此nirS基因常被用于反硝化微生物的检测[11]。自1970年以来,大气中的N2O浓度一直以每年约0.75 μg·L–1的速度增加[12],且全球1/2以上的N2O来自于农田生态系统[1]。因此,系统研究农田±壤的反硝化及其影响机制对调控农田±壤氮循环具有重要的参考意义。
农田±壤往往种植水稻和旱地作物,不同施肥模式下的水田SDP往往高于旱地±壤[13-14],并且影响水田和旱地的微生物机制也可能不同。针对水田±壤,施氮可以明显改变反硝化微生物的群落组成,从而提高水田的SDP[15-16]。已有研究表明,有效磷的缺乏会限制反硝化微生物活性[17-18]。Wei等[19]向缺P水稻±中施入P肥之后,发现通过改变±壤的N/P比,显著降低了amoA基因丰度,增加了反硝化功能基因丰度(narG、nirS、nirK及nosZ)。因此,施P可以通过改变±壤中P的有效性[17-20]和N/P比[21],对±壤反硝化过程产生显著影响。而针对旱地±壤,施氮在短期内也显著改变了旱地±壤中nirS-型反硝化细菌[22]和SDP[23]。Mori等[20]在旱地±壤中加入P之后,发现±壤的硝化及反硝化速率均显著提高。施用K肥能够引起根际±壤中有效钾含量的变化,从而能够影响微生物分泌的酶,改善±壤微域环境,进而调控微生物代谢的一些功能[24-25]。Xue等[26]研究表明:速效钾是影响旱地±壤nirK-型反硝化细菌群落的重要因子。以上研究结果显示,不同种植模式下施肥对±壤反硝化过程的影响机制存在显著差异[27],但是,上述施肥模式对水田和旱地±壤SDP的影响效应及其微生物机制均基于不同地点的水田和旱地获取,不同研究地点存在的±壤母质[28]和气候条件[29]差异可能会影响水田和旱地之间结果的可比性。
本研究选择位于江西进贤的相邻水田和旱地长期定位施肥试验为研究平台,辅以室内培养实验,基于实时荧光定量PCR(qPCR)及高通量测序,开展不同种植模式下施肥对SDP及其与功能基因和微生物群落组成关系的影响研究,主要研究目的包括:(1)明确施肥模式对水田和旱地±壤SDP的不同影响效应;(2)剖析施肥模式对水田和旱地反硝化功能基因narG、nirS、nirK和nosZ丰度,及nirS-型反硝化细菌群落组成影响;(3)评估水田和旱地±壤SDP与功能基因及nirS-型反硝化细菌群落组成之间的潜在关系。以期为探索种植模式与施肥措施对±壤反硝化微生物学机制的双重影响提供参考,为优化农田温室气体减排方案提供依据。
1 材料与方法 1.1 长期试验概况试验地点位于江西省进贤县的红壤研究所(116°20′24″N,28°15′30″E)。水田、旱地均采用随机区组设计,每个处理均3个重复。水田选取4个施肥处理:常规平衡施肥(NPK)、缺钾(NP)、缺磷(NK)和缺氮(PK)处理。旱地的4个施肥处理是:常规平衡施肥(NPK)处理、缺钾(NP)、缺磷(NK)和缺氮钾(P)处理。
1.2 样品采集与处理在2015年12月冬季完成样品采集,每个小区随机选取5点,采集0~20 cm耕层±混匀。取约100 g新鲜±样–80℃保存,用于±壤反硝化微生物的测定;取约50 g新鲜±在4℃条件下贮存,用于铵态氮(NH4+-N)和硝态氮(NO3–-N)的测定;剩余±样在室温下风干,移除肉眼可见的落叶、植物根系及石块,过2 mm筛,用于后期±壤理化性质的测定。
1.3 ±壤理化性质的测定±壤pH按±水比1︰5充分混合后用pH计测定;±壤有机碳(SOC)用重铬酸钾氧化外加热法测定[30]。全氮(TN)用凯氏法测定[30]。NO3–-N和NH4+-N用2 mol·L–1 KCl提取后用流动注射自动分析仪测定[30]。有效磷(AP)用0.5 mol·L–1 NaHCO3提取,钼蓝法测定[30]。±壤速效钾(AK)用CH3COONH4提取[30],火焰光度计测定,检测限为0.004 mmol·L–1。采用草酸铵酸性法提取非晶态氧化铁(Feo)和非晶态氧化铝(Alo)[31],利用ICP-MS测定提取液中的铁和铝。
1.4 厌氧培养试验乙炔抑制厌氧培养的具体过程如下:风干±含水量调节至40% WFPS,25℃黑暗条件中预激活1周。取相当于6 g干±的预激活±至120 mL血清培养瓶中,加入8.658 mg硝酸钾(N 200 μg·g–1干±)和20.508 mg乙酸钠(C 1 mg·g–1干±),调节含水量至70% WFPS。血清培养瓶用黑色橡胶塞和铝盖密封,用真空泵抽真空2 min,注入120 mL纯Ar(99.999%),再进行抽真空处理,此过程重复3次。最后向培养瓶内分别注入108 mL Ar和12 mL C2H2(10% v/v)。25℃条件下,以180 r·min–1振荡15 min,然后在25℃的培养箱中避光培养6 h。用注射器从培养瓶顶部采集气体并储存在20 mL采气瓶中。之后培养瓶抽真空,重新注入Ar,此过程重复3次,以确保培养瓶的厌氧条件,培养瓶再次注入12 mL C2H2和108 mL Ar,进行下一阶段的6 h厌氧培养。培养总共进行24 h。培养期间,N2O-N的平均排放速率代表SDP。
1.5 ±壤反硝化势(N2O-N)的测定气体样品采用安捷伦气相色谱仪(GC,Agilent 7980,Agilent Technologies,Santa Clara,CA,USA)测定。代表SDP的N2O的排放量计算:
| $ F = \frac{{dc}}{{dt}}\frac{N}{{Vm}}V\frac{{273}}{{273 + T}}\frac{1}{M}$ | (1) |
式中,F表示N2O-N的排放速率(N μg·kg–1·h–1),dc/dt表示单位时间内气体浓度的变化量(mg·L–1·h–1),M表示N2O的摩尔质量(28 g·mol–1,N2O-N),Vm表示摩尔体积(22.4 L·mol–1),V表示培养瓶的有效体积(0.12 L),T表示培养温度(25℃),M表示培养±干重(kg)。
1.6 实时荧光定量PCR(qPCR)和测序分析±壤DNA提取采用Fast DNA Spin Kit for Soil(MP Biomedicals,Santa Ana,CA,USA)试剂盒。每个±样连续提取3次,混匀,降低DNA提取偏差。提取的DNA在–80℃条件下保存。采用SYBR Premix Ex TaqTM试剂盒于CFX96 Optical Real-Time PCR System扩增仪(Bio-Rad Laboratories,Hercules,CA)上进行qPCR分析。narG、nirS、nirK及nosZ基因的扩增体系、引物信息及扩增条件参照Wang等[13]。标准曲线用各基因的代表性质粒进行10倍梯度稀释获得,共5个梯度,ddH2O为阴性对照。narG、nirS、nirK和nosZ基因的扩增效率分别为90.4%~93.1%、84.1%~92.8%、86.0%~87.8%和89.8%~89.0%。
利用Illumina MiSeq测序平台(Illumina,San Diego,CA,USA)对nirS基因的PCR扩增产物进行了测序,引物为cd3aF/R3cd。
1.7 数据分析利用SPSS 20.0(Chicago,USA)和R 3.6.1进行数据分析。
2 结果 2.1 ±壤理化性质表 1结果表明,不同施肥模式对水田pH、SOC、TN、NH4+-N和Alo没有显著影响;与NPK相比,NP处理的NO3–-N、C/N和N/P分别降低86.75%、3.51%和96.84%,而AP和Feo含量明显提高346.7%和13.11%;NK处理的AP含量显著降低48.15%,而AK含量和N/P明显提高25.08%、107.37%;PK处理的NO3–-N、C/N和N/P分别下降83.43%、5.15%和96.84%。
|
|
表 1 长期不同施肥模式对水田和旱地土壤基本性质的影响 Table 1 Soil chemical properties of the paddy field and upland relative to fertilization regime |
长期不同施肥模式对旱地TN、NH4+-N及C/N没有显著影响;与平衡施肥NPK相比,NP处理显著提高±壤pH和AP含量,分别提高4.88%和11.36%,而SOC、NO3–-N、AK和N/P分别降低9.97%、75.14%、74.43%和78.05%;NK处理的±壤pH、NO3–-N、AP、Feo和N/P分别降低4.21%、59.53%、55.79%、8.11%和8.54%,Alo含量显著提高;单施P处理,±壤pH和AP显著提高16.35%和43.31%,但SOC、NO3–-N、AK、Feo、Alo和N/P显著降低11.03%、88.34%、56.49%、9.96%、23.63%和91.46%。
2.2 ±壤反硝化势不同施肥模式显著影响水田和旱地的SDP,但影响规律不同。在水田中,NP处理的SDP最高,其次为PK、NPK,NK处理的SDP最低;与NPK相比,NP和PK处理的SDP分别提高33.01%和23.57%,而NK处理则降低35.76%(图 1a)。
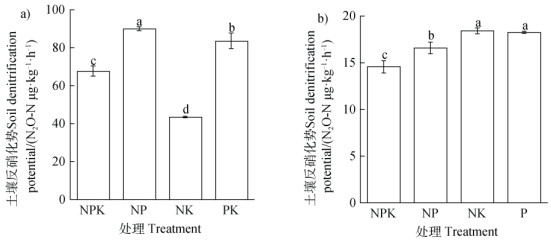
|
注:图中不同小写字母表示不同施肥模式下差异显著,(Duncan多重比较,P<0.05)下同。 Note: Different lowercase letters mean significant difference between different fertilization regimes(Duncan's multiple range test, P<0.05). The same below 图 1 长期不同施肥模式对水田(a)和旱地(b)反硝化势的影响 Fig. 1 Soil denitrification potential in the paddy field(a)and upland(b)relative to fertilization regime |
旱地NK和P处理SDP最高,并且NK和P处理间没有显著差异,其次为NP处理,而NPK处理最低;相对于NPK处理,NK、P和NP处理的SDP分别显著提高26.51%、25.41%和13.94%(图 1b)。
2.3 ±壤反硝化功能基因及与SDP之间关系除了水田的narG基因,本研究采用的施肥模式均显著影响nirS、nirK和nosZ基因的丰度(图 2)。图 2表明,在水田中,与NPK处理相比,NP处理显著提高nirS基因丰度,提高22.94%,而NK和PK处理则显著降低24.07%和19.80%;NK处理的nirK基因丰度显著降低26.08%,NP和PK处理没有显著变化;NP和NK处理的nosZ基因丰度分别显著提高22.45%和17.98%。Spearman相关性分析表明,水田SDP变化与nirS基因丰度显著正相关(r = 0.783**),与narG、nirK和nosZ基因丰度没有显著相关性(表 2)。
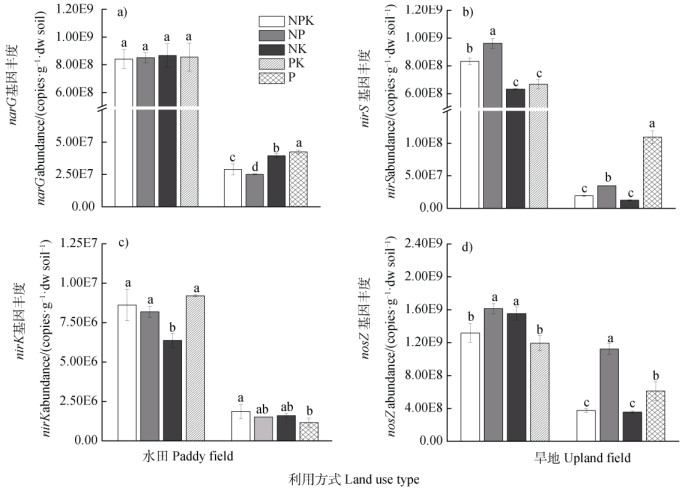
|
图 2 长期不同施肥模式对水田、旱地反硝化功能基因丰度的影响 Fig. 2 Abundances of narG, nirS, nirK and nosZ genes in the paddy field and upland field relative to fertilization regime |
|
|
表 2 水田、旱地反硝化势与功能基因的Spearman相关分析 Table 2 Spearman's correlations analysis of SDPs of the paddy field and upland field with functional genes |
在旱地中(图 2),与NPK处理相比,NP处理的narG基因丰度显著降低12.97%,而NK和P处理显著提高46.90%和46.94%;NK处理对nirS基因丰度没有显著影响,NP和P处理显著提高80.36%和468.19%;nirK基因丰度不受NP和NK处理影响,单施P肥显著降低38.40%;NP和P处理显著提高nosZ基因丰度,分别提高8035%和116.5%。Spearman相关性分析表明,旱地SDP变化与nirG基因丰度显著正相关(r=0.783**),与nirS、nirK和nosZ基因丰度没有显著相关性(表 2)。
2.4 nirS-型反硝化细菌群落组成水田和旱地24个±壤样品测序总共获得415 848条高质量序列,在97%相似度聚类下得到4 727个OTUs。从门水平分析,水田样品的nirS-型反硝化细菌有97.49%来自Proteobacteria,包括Betaproteobacteria(62.39%)、Alphaproteobacteria(30.16%)和Gammaproteobacteria(4.93%);旱地样品中有99.28%来自Proteobacteria,包括Betaproteobacteria(74.85%)、Gammaproteobacteria(19.29%)和Alphaproteobacteria(5.14%)。
置换多元方差分析结果表明,水田nirS-型反硝化细菌群落组成只有PK处理与NPK处理存在显著差异(P < 0.05),而NP和NK处理与NPK处理间均不存在显著差异(表 3);而对旱地而言,仅NP处理的nirS-型反硝化细菌群落组成与NPK处理间无显著差异,NK和P处理与NPK处理间均存在显著差异(P < 0.05)。
|
|
表 3 水田和旱地nirS-型反硝化细菌群落组成的置换多元方差分析 Table 3 PERMANOVA analysis of community composition of nirS-type denitrifier in the paddy filed and upland |
进一步采用火山灰图分析不同施肥处理间nirS-型反硝化细菌的差异,结果表明,在水田中,NK和NP与NPK处理无显著差异的OTUs(图 3a,图 3b),这与表 3结果一致;而PK处理与NPK处理之间微生物差异主要表现为PK处理中的1个OTU(Azospira sp. NC3H-14)丰度显著增加,而11个OTUs丰度显著降低(图 3c)。在旱地中,NK与NPK处理之间的差异主要表现为NK处理具有更高丰度的3个OTUs,它们归类为Azospira sp. NC3H-14和Ideonella sp. NC3L-43b(图 3e)。而P与NPK处理间的主要差异表现为P处理中具有更高丰度的72个OTUs,它们主要归类为Azospira sp. NC3H-14、Rhodanobacter sp. D206a和Rubrivivax gelatinosus(图 3f)。

|
注:黑点代表丰富型物种,灰点代表消弭型物种,空心点代表稳定型物种。(a)、(b)、(c)分别是红壤水田NP、NK、PK与NPK对比,(d)、(e)、(f)分别是红壤旱地NP、NK、P与NPK对比。 Note: Black point represents OTUs enriched, gray point represents OTUs depleted, and the hollow point represents the stable OTUs.(a), (b), and(c) stands for NP vs. NPK, NK vs. NPK, and PK vs. NPK in the paddy field, respectively.(d), (e), and(f) are NP vs. NPK, NK vs. NPK, and P vs. NPK in the upland field, respectively. 图 3 长期不同施肥模式下水田和旱地OTU水平nirS-型反硝化细菌差异物种 Fig. 3 Volcano plots showing enrichment(black) and depletion(gray) of nirS-type denitrifiers at the OTU level in the paddy field and upland relative to treatment |
反硝化功能基因丰度对SDP的影响一直以来存在不确定性。有研究认为反硝化功能基因丰度与SDP没有直接的关系[32],也有研究指出反硝化功能基因丰度变化能显著影响SDP[6,33]。本研究通过Spearman相关分析研究发现水田SDP变化与nirS基因丰度显著正相关(r=0.783**),而与narG、nirK和nosZ基因丰度没有显著相关性(表 2),这与罗希茜等[34]的研究结果一致,其研究结果表明施肥导致的水田SDP变化与nirS基因丰度有关。与NPK处理相比,NP处理同时提高了nirS基因丰度和SDP,而NK处理同时降低了nirS基因丰度和SDP。上述结果表明,与NPK处理相比,NP处理和NK处理的SDP差异与nirS基因丰度的变化有关。±壤微生物生物量通常较±壤和植物具有更低的N/P,微生物更容易受到磷的限制[35]。Mori等[20]及Hartman和Richardson[36]通过对缺磷±壤反硝化的研究发现,±壤AP含量的提高可以促进±壤氮循环及反硝化微生物的生长,从而提高±壤反硝化速率。Wei等[19]研究也发现,通过改变±壤的C/P和N/P,可以显著提高反硝化功能基因丰度。因此推断NP处理中较高的AP含量以及较低的N/P可能会引起nirS基因丰度的增加,使其SDP显著高于平衡施肥NPK。而NK处理中较低的AP含量以及较高的N/P可能降低了nirS基因丰度,从而可能与NK处理较低的SDP有关(图 1)。
与水田不同,不同施肥模式下,旱地SDP变化与narG基因丰度显著正相关(r=0.592*)(表 2),这与Dong等[37]的研究结果非常相似,其研究发现narG基因丰度显著影响玉米-小麦生态系统中N2O的排放。与NPK处理相比,P和NK处理同时提高了narG基因丰度和SDP,表明narG基因丰度的变化与P和NK处理的SDP差异有关。不同施肥模式下,NPK和NP处理的Feo含量明显高于NK和P处理。Feo胶体对阴阳离子的吸附性限制反应基质的有效性,例如SOC的有效性,从而降低narG基因丰度[38]。因此,NK和P处理中较低的Feo可能引起narG基因丰度显著高于NPK处理,从而使P和NK处理具有较高的SDP。
3.2 水田和旱地影响SDP与nirS-型反硝化细菌群落组成的关系在水田中,Azospira sp. NC3H-14是PK和NPK处理间具有显著差异的反硝化微生物,而NP和NK处理与NPK处理间反硝化细菌群落组成间没有差异(图 3c)。Azospira菌属不能利用碳水化合物,但可利用有机酸、氨基酸等为碳源,氧或硝酸盐作为末端电子受体[39]。Li等[40]研究发现,与NPK相比,不平衡施肥改变±壤有机碳结构,且养分有效性较养分含量对微生物的影响更大[41]。因此,PK施肥可能改变了±壤有机碳结构使其更容易被Azospira NC3H-14利用,而提高了Azospira sp. NC3H-14的丰度,并提高了PK施肥处理的SDP。
在旱地中,与NPK处理相比,NK处理的Azospira sp. NC3H-14和Ideonella sp. NC3L-43b丰度更高,而P处理的Azospira sp. NC3H-14、Rhodanobacter sp. D206a和Rubrivivax gelatinosus具有更高的丰度(图 3)。有研究表明,在厌氧条件下,Rubrivivax gelatinosus可以通过将亚硝酸盐还原为N2来提高SDP[42-43]。上述反硝化细菌丰度的增加与NK及P处理显著改变nirS-型反硝化细菌群落组成有关(表 3),进而提高NK和P处理的SDP。旱地NP处理SDP的变化与本研究测定的功能基因丰度和nirS-型反硝化细菌组成均无关,这可能与其他本研究未测定的参数变化有关。比如,反硝化酶活性也是调控SDP变化的重要因素[44-45]。此外,Chee-Sanford等[46]研究发现,非特异性nosZ基因(Clade Ⅱ nosZ)较特异性扩增的nosZ基因微生物在生态系统中(特别是±壤中)更具有优势。因此,非特异性nosZ基因在水田、旱地不同种植模式下可能会对SDP有不同的影响。而本研究中的nosZ基因扩增引物无法反应非特异性nosZ基因代表的微生物。因此,影响水田和旱地间SDP变化的机理值得进一步深入研究。
4 结论通过直接比较相同±壤母质和气候条件下的水田和旱地SDP变化发现,与常规平衡施肥NPK相比,水田NP和PK处理的SDP显著提高,而NK处理显著降低,其中NP和NK的SDP变化主要与nirS基因丰度变化有关,而PK处理较高的SDP与反硝化细菌Azospira sp. NC3H-14的丰度增加有关;而在旱地中,NP、NK、P处理的SDP均显著提高,其中NK和P处理的SDP变化与narG基因丰度显著正相关,同时与NK处理中反硝化细菌Azospira sp. NC3H-14和Ideonella sp. NC3L-43b丰度显著增加,P处理中Azospira sp. NC3H-14、Rhodanobacter sp. D206a和Rubrivivax gelatinosus丰度显著增加有关。
| [1] |
Hu H W, Chen D L, He J Z. Microbial regulation of terrestrial nitrous oxide formation: Understanding the biological pathways for prediction of emission rates[J]. FEMS Microbiology Reviews, 2015, 39(5): 729-749. DOI:10.1093/femsre/fuv021
(  0) 0) |
| [2] |
Wrage N, Velthof G L, van Beusichem M L, et al. Role of nitrifier denitrification in the production of nitrous oxide[J]. Soil Biology & Biochemistry, 2001, 33(12/13): 1723-1732.
(  0) 0) |
| [3] |
Philippot L, Hallin S, Schloter M. Ecology of denitrifying prokaryotes in agricultural soil[J]. Advances in Agronomy, 2007, 96: 249-305.
(  0) 0) |
| [4] |
Sun R B, Guo X S, Wang D Z, et al. Effects of long-term application of chemical and organic fertilizers on the abundance of microbial communities involved in the nitrogen cycle[J]. Applied Soil Ecology, 2015, 95: 171-178. DOI:10.1016/j.apsoil.2015.06.010
(  0) 0) |
| [5] |
Wolsing M, Priemé A. Observation of high seasonal variation in community structure of denitrifying bacteria in arable soil receiving artificial fertilizer and cattle manure by determining T-RFLP of nir gene fragments[J]. FEMS Microbiology Ecology, 2004, 48(2): 261-271. DOI:10.1016/j.femsec.2004.02.002
(  0) 0) |
| [6] |
Chen Z, Hou H J, Zheng Y, et al. Influence of fertilisation regimes on a nosZ-containing denitrifying community in a rice paddy soil[J]. Journal of the Science of Food and Agriculture, 2012, 92(5): 1064-1072. DOI:10.1002/jsfa.4533
(  0) 0) |
| [7] |
Buermans H P J, den Dunnen J T. Next generation sequencing technology: Advances and applications[J]. Biochimica et Biophysica Acta: Molecular Basis of Disease, 2014, 1842(10): 1932-1941. DOI:10.1016/j.bbadis.2014.06.015
(  0) 0) |
| [8] |
Cutruzzola F, Brown K, Wilson E K, et al. The nitrite reductase from Pseudomonas aeruginosa: Essential role of two active-site histidines in the catalytic and structural properties[J]. Proceedings of the National Academy of Sciences of the United States of America, 2001, 98(5): 2232-2237. DOI:10.1073/pnas.041365298
(  0) 0) |
| [9] |
Zumft W G. Cell biology and molecular basis of denitrification[J]. Microbiology and Molecular Biology Reviews, 1997, 61(4): 533-616.
(  0) 0) |
| [10] |
Throbäck I N, Enwall K, Jarvis Å, et al. Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE[J]. FEMS Microbiology Ecology, 2004, 49(3): 401-417. DOI:10.1016/j.femsec.2004.04.011
(  0) 0) |
| [11] |
Braker G, Fesefeldt A, Witzel K P. Development of PCR primer systems for amplification of nitrite reductase genes(nirK and nirS) to detect denitrifying bacteria in environmental samples[J]. Applied and Environmental Microbiology, 1998, 64(10): 3769-3775. DOI:10.1128/AEM.64.10.3769-3775.1998
(  0) 0) |
| [12] |
IPCC. Climate change 2014: Mitigation of climate change, summary for policymakers[M]//Contribution of working group Ⅲ to the fifth assessment report of the intergovernmental panel on climate change. Cambridge: Cambridge University Press, 2014.
(  0) 0) |
| [13] |
Wang H C, Liu Z D, Ma L, et al. Denitrification potential of paddy and upland soils derived from the same parent material respond differently to long-term fertilization[J]. Frontiers in Environmental Science, 2020, 8: 105. DOI:10.3389/fenvs.2020.00105
(  0) 0) |
| [14] |
Xu Y B, Cai Z C. Denitrification characteristics of subtropical soils in China affected by soil parent material and land use[J]. European Journal of Soil Science, 2007, 58(6): 1293-1303. DOI:10.1111/j.1365-2389.2007.00923.x
(  0) 0) |
| [15] |
Wang L, Sheng R, Yang H C, et al. Stimulatory effect of exogenous nitrate on soil denitrifiers and denitrifying activities in submerged paddy soil[J]. Geoderma, 2017, 286: 64-72. DOI:10.1016/j.geoderma.2016.10.023
(  0) 0) |
| [16] |
Wang J, Cheng Y, Cai Z C, et al. Effects of long-term fertilization on key processes of soil nitrogen cycling in agricultural soil: A review (In Chinese)[J]. Acta Pedologica Sinica, 2016, 53(2): 292-304. [王敬, 程谊, 蔡祖聪, 等. 长期施肥对农田土壤氮素关键转化过程的影响[J]. ±壤学报, 2016, 53(2): 292-304.]
(  0) 0) |
| [17] |
He M Z, Dijkstra F A. Phosphorus addition enhances loss of nitrogen in a phosphorus-poor soil[J]. Soil Biology & Biochemistry, 2015, 82: 99-106.
(  0) 0) |
| [18] |
Sheng R, Meng D L, Wu M N, et al. Effect of agricultural land use change on community composition of bacteria and ammonia oxidizers[J]. Journal of Soils and Sediments, 2013, 13(7): 1246-1256. DOI:10.1007/s11368-013-0713-3
(  0) 0) |
| [19] |
Wei X M, Hu Y J, Peng P Q, et al. Effect of P stoichiometry on the abundance of nitrogen-cycle genes in phosphorus-limited paddy soil[J]. Biology and Fertility of Soils, 2017, 53(7): 767-776. DOI:10.1007/s00374-017-1221-1
(  0) 0) |
| [20] |
Mori T, Ohta S, Ishizuka S, et al. Effects of phosphorus addition on N2O and NO emissions from soils of an Acacia mangium plantation[J]. Soil Science and Plant Nutrition, 2010, 56(5): 782-788. DOI:10.1111/j.1747-0765.2010.00501.x
(  0) 0) |
| [21] |
Zhang Y, Song C L, Ji L, et al. Cause and effect of N/P ratio decline with eutrophication aggravation in shallow lakes[J]. Science of the Total Environment, 2018, 627: 1294-1302. DOI:10.1016/j.scitotenv.2018.01.327
(  0) 0) |
| [22] |
Mo X H, Ma W, Shi R J, et al. Diversity of nirS-type denitrifying bacteria under different nitrogen fertilizer management in wheat soil (In Chinese)[J]. Acta Microbiologica Sinica, 2009, 49(9): 1203-1208. DOI:10.3321/j.issn:0001-6209.2009.09.012 [莫旭华, 麻威, 史荣久, 等. 氮肥对小麦田±壤nirS型反硝化细菌多样性的影响[J]. 微生物学报, 2009, 49(9): 1203-1208.]
(  0) 0) |
| [23] |
Wang J, Yan X. Denitrification in upland of China: Magnitude and influencing factors[J]. Journal of Geophysical Research: Biogeosciences, 2016, 121(12): 3060-3071. DOI:10.1002/2016JG003541
(  0) 0) |
| [24] |
田悦悦. 长期缺钾对设施番茄根系分泌物及根际微生态的影响[D]. 沈阳: 沈阳农业大学, 2018. Tian Y Y. Effects of long-term potassium deficiency on tomato root exudates and rhizosphere microecology[D]. Shenyang: Shenyang Agricultural University, 2018. (  0) 0) |
| [25] |
Wang Y Y, Lu S E, Chen X M, et al. Analyzing the nitrate reductase gene(nirK) community in the peat soil of the Zoige Wetland of the Tibetan Plateau (In Chinese)[J]. Acta Ecologica Sinica, 2017, 37(19): 6607-6615. [王蓥燕, 卢圣鄂, 陈小敏, 等. 若尔盖高原湿地泥炭沼泽±亚硝酸盐还原酶(nirK)反硝化细菌群落结构分析[J]. 生态学报, 2017, 37(19): 6607-6615.]
(  0) 0) |
| [26] |
Xue K, Wu L Y, Deng Y, et al. Functional gene differences in soil microbial communities from conventional, low-input, and organic farmlands[J]. Applied and Environmental Microbiology, 2013, 79(4): 1284-1292. DOI:10.1128/AEM.03393-12
(  0) 0) |
| [27] |
Zulkarnaen N, Cheng Y, Zhang J B. Denitrification potential and gas emission in red soils under different land use types (In Chinese)[J]. Soils, 2020, 52(2): 348-355. [Nanang Zulkarnaen, 程谊, 张金波. 不同利用方式红壤反硝化势和气态产物排放特征[J]. ±壤, 2020, 52(2): 348-355.]
(  0) 0) |
| [28] |
Xing X Y, Sheng R, Xu H F, et al. Denitrification characteristics of dryland soils derived from different parent materials (In Chinese)[J]. Soils, 2019, 51(5): 949-954. [邢肖毅, 盛荣, 徐慧芳, 等. 不同母质发育旱地±壤反硝化功能差异及其关键影响因素[J]. ±壤, 2019, 51(5): 949-954.]
(  0) 0) |
| [29] |
Wang J Y, Chadwick D R, Cheng Y, et al. Global analysis of agricultural soil denitrification in response to fertilizer nitrogen[J]. Science of the Total Environment, 2018, 616/617: 908-917. DOI:10.1016/j.scitotenv.2017.10.229
(  0) 0) |
| [30] |
Page A L, Miller R H, Keeney D R. Methods of soil analysis: Chemical and microbiological properties . Madison, Wisconsin: Soil Science Society of America, 1982.
(  0) 0) |
| [31] |
Pansu M, Gautheyrou J. Handbook of soil analysis: Mineralogical, organic and inorganic methods groenekennis . Berlin, Heidelberg, New York: Springer, 2006.
(  0) 0) |
| [32] |
Liu Y, Shen K, Wu Y C, et al. Abundance and structure composition of nirK and nosZ genes as well as denitrifying activity in heavy metal-polluted paddy soils[J]. Geomicrobiology Journal, 2018, 35(2): 100-107. DOI:10.1080/01490451.2017.1333175
(  0) 0) |
| [33] |
Cui P Y, Fan F L, Yin C, et al. Long-term organic and inorganic fertilization alters temperature sensitivity of potential N2O emissions and associated microbes[J]. Soil Biology & Biochemistry, 2016, 93: 131-141.
(  0) 0) |
| [34] |
Luo X Q, Chen Z, Hu R G, et al. Effect of long-term fertilization on the diversity of nitrite reductase genes(nirK and nirS) in paddy soil (In Chinese)[J]. Environmental Science, 2010, 31(2): 423-430. [罗希茜, 陈哲, 胡荣桂, 等. 长期施用氮肥对水稻±亚硝酸还原酶基因多样性的影响[J]. 环境科学, 2010, 31(2): 423-430.]
(  0) 0) |
| [35] |
Cleveland C C, Liptzin D. C: N: P stoichiometry in soil: Is there a "Redfield ratio" for the microbial biomass?[J]. Biogeochemistry, 2007, 85(3): 235-252. DOI:10.1007/s10533-007-9132-0
(  0) 0) |
| [36] |
Hartman W H, Richardson C J. Differential nutrient limitation of soil microbial biomass and metabolic quotients(qCO2): Is there a biological stoichiometry of soil microbes?[J]. PLoS One, 2013, 8(3): e57127. DOI:10.1371/journal.pone.0057127
(  0) 0) |
| [37] |
Dong Z X, Zhu B, Jiang Y, et al. Seasonal N2O emissions respond differently to environmental and microbial factors after fertilization in wheat-maize agroecosystem[J]. Nutrient Cycling in Agroecosystems, 2018, 112(2): 215-229. DOI:10.1007/s10705-018-9940-8
(  0) 0) |
| [38] |
Turner S, Mikutta R, Guggenberger G, et al. Distinct pattern of nitrogen functional gene abundances in top-and subsoils along a 120, 000-year ecosystem development gradient[J]. Soil Biology & Biochemistry, 2019, 132: 111-119.
(  0) 0) |
| [39] |
Reinhold-Hurek B, Hurek T. The genera Azoarcus, Azovibrio, Azospira and Azonexus[M]//The Prokaryotes. New York: Springer, 2006: 873-891.
(  0) 0) |
| [40] |
Li D D, Chen L, Xu J S, et al. Chemical nature of soil organic carbon under different long-term fertilization regimes is coupled with changes in the bacterial community composition in a Calcaric Fluvisol[J]. Biology and Fertility of Soils, 2018, 54(8): 999-1012. DOI:10.1007/s00374-018-1319-0
(  0) 0) |
| [41] |
Briar S S, Fonte S J, Park I, et al. The distribution of nematodes and soil microbial communities across soil aggregate fractions and farm management systems[J]. Soil Biology & Biochemistry, 2011, 43(5): 905-914.
(  0) 0) |
| [42] |
Magoč T, Salzberg S L. FLASH: fast length adjustment of short reads to improve genome assemblies[J]. Bioinformatics, 2011, 27(21): 2957-2963. DOI:10.1093/bioinformatics/btr507
(  0) 0) |
| [43] |
Edwards J, Johnson C, Santos-Medellín C, et al. Structure, variation, and assembly of the root-associated microbiomes of rice[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(8): E911-E920. DOI:10.1073/pnas.1414592112
(  0) 0) |
| [44] |
Shi Y, Huang G H. Relationship between soil denitrifying enzyme activities and N2 O emission (In Chinese)[J]. Chinese Journal of Applied Ecology, 1999, 10(3): 329-331. DOI:10.3321/j.issn:1001-9332.1999.03.019 [史奕, 黄国宏. ±壤中反硝化酶活性变化与N2O排放的关系[J]. 应用生态学报, 1999, 10(3): 329-331.]
(  0) 0) |
| [45] |
Wang L F, Cai Z C. Effects of temperature and water regime on nitrification and denitrification activity of upland red soils (In Chinese)[J]. Soils, 2004, 36(5): 543-546, 560. [王连峰, 蔡祖聪. 水分和温度对旱地红壤硝化活力和反硝化活力的影响[J]. 土壤, 2004, 36(5): 543-546, 560.]
(  0) 0) |
| [46] |
Chee-Sanford J C, Connor L, Krichels A, et al. Hierarchical detection of diverse Clade Ⅱ(atypical) nosZ genes using new primer sets for classical-and multiplex PCR array applications[J]. Journal of Microbiological Methods, 2020, 172: 105908. DOI:10.1016/j.mimet.2020.105908
(  0) 0) |
 2022, Vol. 59
2022, Vol. 59


