2. 中国科学院大学, 北京 100049
2. University of Chinese Academy of Sciences, Beijing 100049, China
土壤微生物所介导的氮素循环在调控生物地球化学循环和维持生态系统功能方面起着重要的作用。土壤微生物参与了包括固氮、氨化、硝化和反硝化等重要生态过程,氮循环中的微生物群落组成和功能会影响氮的生物利用度,进而调控作物的生长特征。此外,反硝化和氨氧化过程是温室气体N2O排放的主要原因,其对温室效应的产生大约是CO2的300倍[1]。因此对氮循环微生物的空间分布特征研究,不仅是微生物演变和进化的基础科学问题,也是预测微生物及其所介导的生态功能对环境条件变化响应、适应和反馈的理论依据。
关于微生物空间分布研究,目前已经排除了随机分布格局的假说,研究的重点转为当代环境因子和历史进化因素驱动土壤微生物空间分布的内在机制上[2]。已有的研究表明,微生物群落受到环境和历史因素的双重制约[3-4],而地理分割对微生物群落分布的贡献与采样的空间尺度有关[5-7]。Jiang等[8]针对未受干扰的湿地土壤的研究结果表明,除环境因素外,地理距离对反硝化微生物群落的分布特征具有重要影响。Hu等[9]基于空间尺度对氨氧化微生物的分布规律研究发现,土壤pH而非地理距离是影响氨氧化微生物分布的重要因子。此外,Yao等[10]在苏格兰全区域针对氨氧化微生物的群落分布研究发现,包括土地利用方式、土壤类型、气候条件和氮沉降等诸多因子协同驱动了氨氧化微生物的空间分布。然而,目前关于氮循环研究多基于单一的氮循环过程,而关于氮循环关键循环过程微生物空间分布模式的关联分析相对较少[11-12]。
东北黑土地是我国重要的优质商品粮基地,提供了占全国33%的调出商品粮,直接关系着国家的粮食生产和生态安全。东北黑土区长期的过度垦殖、重用轻养等不合理的耕作方式,导致了黑土区生物丰度和生态功能稳定性降低,黑土耕地质量退化和土壤地力下降等问题。东北黑土区从南到北年平均气温逐渐下降,但土壤全C、N和P含量,土壤酶活性和微生物量呈上升趋势[13-14]。东北黑土农田土壤养分含量,特别是土壤有机质含量,呈现南低北高的规律性地理分布特征[13]。因此,相同的土壤类型和有机质的规律性变化特征,使其成为研究土壤微生物生物地理分布规律的理想区域。已有研究证实,东北黑土区农田细菌和真菌群落结构存在明显的地理分布格局[15-16],但关于氮循环主要功能微生物的分布规律和驱动机制目前还尚不清楚。本研究采用实时荧光定量PCR技术,对氮循环关键过程中的固氮、硝化和反硝化特异基因丰度进行耦联分析,旨在解析东北黑土区氮循环关键过程微生物基因丰度变化的异同关系,揭示氮循环功能微生物基因的空间分布特征及对不同环境条件的响应机制,对提高土壤氮素利用率以及降低生态环境负效应具有重要意义。
1 材料与方法 1.1 土壤样本采集沿东北黑土区,南起辽宁昌图(42°50′N,124°07′E)北至黑龙江嫩江(49°08′N,125°37′E),于2012年9月玉米和大豆的成熟期,选取26个不同有机质含量的旱地农田土壤样本(图 1;[15])。采用对角线取样法,在每个10 m×10 m的样方内,采集10个耕层土壤(0~20 cm)样品,土壤样品均匀混合后过2 mm土筛,置于冰盒中,到达实验室后于–80℃冰箱保存,低温冷冻样品用于土壤DNA提取及下游基因定量分析。余下的土壤风干后,用于土壤基础理化性质测定。供试样点土壤pH变化为4.56至6.57,土壤全碳含量为11.77 g·kg–1至53.53 g·kg–1,土壤pH与纬度没有相关性(P = 0.985),而土壤全碳(P<0.001)与全氮含量(P = 0.003)与纬度存在显著相关性(表 1;[15])。

|
图 1 东北黑土区农田土壤氮循环基因分布研究样点采集图[15] Fig. 1 Sampling map of sites for research on the distribution of soil nitrogen cycling genes in farmland in the black soil zone of Northeast China |
|
|
表 1 土壤样本的地理信息及土壤理化性质[15] Table 1 Geographical information of soil samples and soil physical and chemical properties |
利用pH计测定振荡30 min后的土壤水悬浮液(1:5 w/v),计算土壤pH。采用元素分析仪(VarioEL III,Germany)测定土壤全碳(TC)和全氮(TN)含量。土壤速效钾(AK)利用火焰光度计(ICPS- 7500,Shimadzu,Japan)进行测定。土壤全磷(TP)、速效磷(AP)、铵态氮(NH4+-N)和硝态氮(NO3–-N)利用连续流动分析系统(SKALAR SAN++,Netherlands)进行测定。
1.3 土壤DNA的提取0.5 g均匀土壤样品,利用Fast DNA® Spin Kit For Soil(MP Biomedicals,USA),并按步骤对土壤微生物总DNA进行提取。提取后的DNA溶解在TE缓冲液(10 mmol·L–1 Tris-HCl,1 mmol·L–1 EDTA,pH = 8.0)中,并用NanoDrop® 2000(Thermo Scientific,USA)测定其浓度及质量,保存于–20℃冰箱中备用。
1.4 实时荧光定量PCR分析本研究采用实时荧光定量PCR(qPCR)的方法,确定固氮过程(nifH)、氨氧化过程(氨氧化古菌amoA和氨氧化细菌amoA,即AOA amoA和AOB amoA)、反硝化过程(nirS,nirK和nosZ)的6个功能基因的丰度。qPCR反应在Light Cycler® 480 thermocycler(Roche Applied Science)平台上进行,反应条件和所需的引物见表 2。反应体系共20 µL,包括SYBR® Premix Ex Taq(TaKaRa,Dalian,China)10 µL、正向和反向引物各1 µL(10 µmol·L–1)、土壤样品DNA 1 µL(10 ng·µL–1)、1% BSA(TaKaRa,Dalian,China)0.2 µL(防止腐殖质干扰)和灭菌的去离子水6.8 µL。标准曲线由已插入目标基因片段且拷贝数已知的质粒DNA经10倍梯度稀释而成,每个标准样品进行三次重复。质粒的制作方法如下:采用相应的引物(表 2)对氮循环功能基因进行PCR扩增,PCR产物中的目的基因由胶回收试剂盒(TaKaRa,Dalian,China)进行回收纯化。然后将纯化后的目的基因连接到质粒载体(pMD18-T)上,并导入至感受态细胞中。经蓝白斑筛选,挑出阳性克隆摇菌培养24 h,部分菌液送至华大基因(Shenzhen,China)进行测序鉴定以确保质粒中插入片段即为目的基因,其余菌液用质粒提取试剂盒(TaKaRa,Dalian,China)提取质粒DNA。最后,用Qubit fluorometer(Invitrogen,USA)测定质粒浓度,并计算质粒拷贝数:质粒拷贝数(copies·µL–1)= 6.02 × 1023 ×质粒DNA的浓度(ng·µL–1)× 10–9 /[(载体序列长度+目的基因序列长度)× 660]。根据标准曲线的回归方程(R2 =0.993 0 ~ 0.999 4),通过Ct(Threshold cycle)值来计算土壤样品中相应基因的拷贝数。本研究氮循环功能基因qPCR的扩增效率在98%~103%之间,溶解曲线均为单一峰。
|
|
表 2 氮循环功能基因实时荧光定量PCR引物及反应条件 Table 2 Quantitative real-time PCR primers and reaction conditions for nitrogen cycle functional genes |
利用SPSS 16.0软件,基于Duncan多重比较法对不同土壤pH组间氮循环微生物功能基因的丰度进行单因素方差分析。采用R(v.3.3.1)软件,通过“ggplot2”的相关性分析,探究土壤pH和TC含量与氮循环微生物功能基因丰度之间的相关关系。基于Bray-Curtis距离进行非度量多维尺度分析(Non-metric Multidimensional Scaling,NMDS),将NMDS的氮循环基因分布关系与土壤pH和TC进行回归分析,以解析氮循环微生物基因丰度组成和环境因子间的关系。此外,用R软件的“vegan”和“randomForest”分别进行Mantel test分析、方差分解分析(Variance Partitioning Analysis,VPA)以及随机森林分析(Random Forest,RF),以阐明东北黑土区氮循环关键过程微生物空间分布格局的主要土壤驱动因子。
2 结果 2.1 氮循环微生物功能基因丰度分布基于qPCR分析黑土区农田土壤氮循环关键过程微生物功能基因丰度(拷贝数·g–1干土)的结果显示,固氮微生物nifH基因丰度为4.57×105(德惠)至6.04×106(北安);反硝化微生物nirK基因丰度为2.51×105(德惠)至1.38×106(北安)、nirS基因丰度为3.02×105(德惠)至2.97×106(北安),而nosZ基因丰度为7.18×106(德惠)至4.70×107(北安);硝化微生物AOA amoA基因丰度为2.51×106(德惠)至3.88×107(北安),而AOB amoA基因丰度为2.92×104(德惠)至2.19×106(巴彦)。研究发现,在低pH(4.5~5.0)的土壤中,不同氮循环基因丰度均显著低于其他pH土壤样本,而pH > 6.0样本中不同土壤氮循环基因丰度最高;种植大豆的土壤(哈尔滨)nifH基因丰度显著高于种植玉米(呼兰、双城等)的土壤样本(分别高于60%和83%);AOA amoA基因丰度显著高于AOB amoA基因丰度,AOA amoA与AOB amoA的丰度比值为3.1至91.0之间。相同采样点间不同反硝化过程nirS数量显著高于nirK(高于15%~235%)(图 2)。
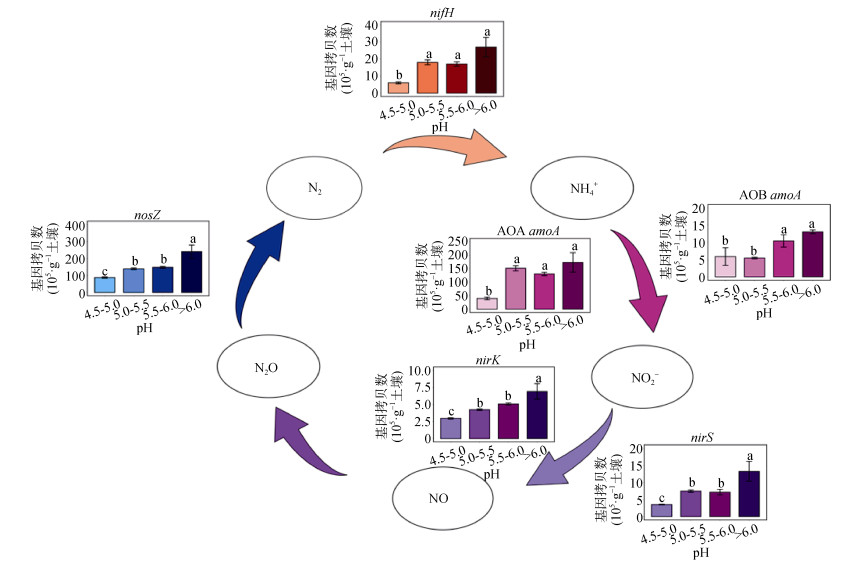
|
注:用Duncan法统计(n = 4),同一基因不同pH之间不同字母表示差异显著(P < 0.05)。 Note: By Duncan method(n=4), different letters between different pH values of the same gene indicate significance level at P < 0.05. 图 2 黑土区不同氮循环功能基因丰度按照土壤pH分组的分布特征 Fig. 2 Abundance of different nitrogen cycling functional genes, and in soils separated according to pH categories in the black soil zone |
氮循环关键过程微生物丰度与土壤因子的相关性分析结果显示,代表不同氮循环过程的6个基因,除AOB amoA与TC没有相关性外(R2 = 0.029,P = 0.14),反硝化过程nirS、nirK和nosZ基因,固氮nifH基因和AOA amoA基因均与土壤pH和TC含量显著正相关(P < 0.001)(图 3)。此外,除土壤pH和TC外,上述功能基因丰度还与纬度和TN含量等土壤因子显著相关,如nifH、AOA amoA和nosZ基因与纬度梯度显著正相关(P < 0.001),nifH、AOA amoA、nirS和nosZ基因分别与TN和TP含量显著正相关(P < 0.001),而AOB amoA基因丰度则与AK、AP、NH4+-N和NO3–-N含量显著正相关(P < 0.001)。
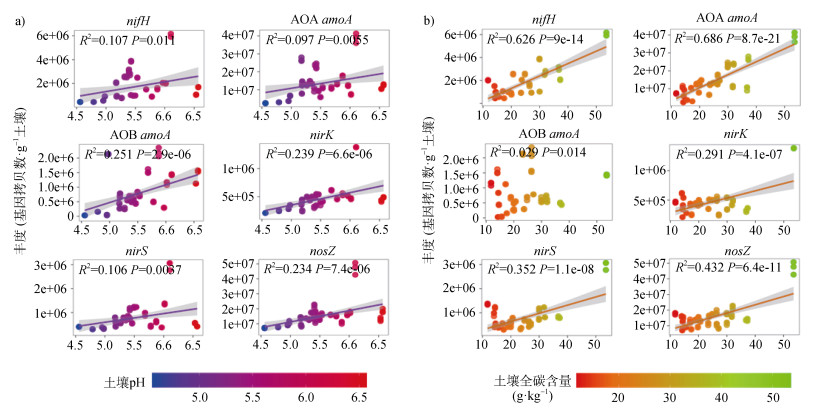
|
图 3 土壤pH(a)和土壤TC(b)含量与不同氮循环功能基因丰度间的相关性分析 Fig. 3 The relationship between soil pH(a)and soil TC(b)and the abundance of the functional genes in different nitrogen cycling of black soil zone |
主要氮循环关键过程微生物丰度的NMDS分析结果显示,氮循环微生物丰度并非随机分布,其分布主要受到土壤pH、TC含量的影响。NMDS1坐标轴分别与土壤pH(r = 0.546,P = 0.004)和TC(r = 0.773,P < 0.001)显著正相关(图 4)。此外,Mantel test分析结果显示除土壤pH和TC外,氮循环微生物基因丰度组成与纬度(r = 0.135,P = 0.049)、TN(r = 0.135,P = 0.021)和TP(r = 0.135,p = 0.021)含量显著相关(表 3)。

|
图 4 农田土壤氮循环关键过程微生物基因丰度组成分布特征,采样点按照pH(a)和TC(b)着色 Fig. 4 The biogeography distribution pattern of soil microbial gene composition related to key processes of nitrogen cycling in agricultural black soils, sites have been colour-coded to soil pH gradient(a)and soil total carbon gradient(b) |
|
|
表 3 黑土区氮循环微生物基因丰度组成与环境因子的Mantel test分析 Table 3 Mantel test analysis between the microbial gene composition involved in nitrogen cycling and environmental factors in the black soil zone |
环境因子对氮循环微生物丰度分布相对贡献的VPA分析结果显示,土壤因子对氮循环微生物丰度分布的总解释度为70.98%,其中土壤pH和TC对氮循环微生物群落分布的解释率分别为6.69%和4.38%,而纬度对氮循环微生物群落分异的解释率为3.76%(图 5a)。此外,基于上述因子的RF分析结果显示,土壤pH和TC与氮循环关键过程微生物基因丰度组成分布显著相关,且是土壤氮循环微生物空间分布的最主要影响因子(图 5b)。

|
注:∗表示显著相关(P < 0.05)。 Note: ∗ indicate significance level at P < 0.05. 图 5 黑土区氮循环微生物基因丰度组成与土壤因子的方差分解分析(a)和随机森林分析(b) Fig. 5 Variance partitioning analysis(a)and random forest analysis(b)between the microbial gene composition involved in nitrogen cycling and environmental factors in the black soil zone |
本研究发现东北黑土区农田土壤pH呈酸性(pH变化范围在4.56~6.57之间),而酸性的土壤环境显著降低了不同氮循环过程微生物基因丰度(图 2)。种植大豆土壤中的nifH基因丰度显著高于种植玉米的土壤,这可能是由于固氮菌与大豆根系形成了共生体,在促进植物生长的同时为固氮菌提供了独特的生态位[12]。以往研究发现AOA与AOB数量的主导性在不同生态系统中存在差异,如Wuchter等[23]和He等[24]发现AOA广泛存在于海洋、湖泊及陆地生态系统中,而且数量通常远远高于AOB。然而,Di等[25]针对草原土壤的研究却得到了相反的研究结果,尽管古菌AOA amoA基因大量存在或相当高的丰度,并不意味着AOA发挥的作用大于AOB。基于长期定位实验,研究发现在酸性土壤中,土壤的硝化作用主要由AOA驱动,一些AOA类群仅出现在强酸性的土壤中[26-27],而在中性或是碱性土壤中,AOB可能更活跃[25,28]。基于空间尺度本研究发现,东北黑土农田土壤中AOA的数量显著高于AOB,证实了酸性土壤中AOA较AOB的amoA基因更具有优势。此外,有研究发现在酸性土壤中作为氨单加氧酶直接底物的NH3电离成NH4+,NH3浓度相对较低,此时对底物的亲和力较高的AOA在氨氧化过程中起主导作用[29]。
参与反硝化过程携带nirK和nirS基因的细菌存在不同的进化史,但却执行相同的生物学功能。以往基于qPCR对nirK和nirS基因丰度的研究结果显示,不同生态系统中nirK基因要显著低于nirS基因丰度,说明环境中携带nirK基因的反硝化细菌可能存在低表达和多样性相对较低的现象[30]。然而,对可培养反硝化细菌nirK和nirS基因的研究发现,这两个基因可能存在水平转移,nirS所编码的蛋白多具有统一分子结构,而nirK编码的蛋白存在几个结构亚类,说明环境中nirK的基因丰度和多样性可能高于nirS基因[31-32]。本研究中相同样点的nirS基因丰度远高于nirK基因的结果,很可能是由于PCR对两类基因扩增的偏好性所导致。Helen等[33]研究发现,利用PCR扩增的方式仅能检测到I-type nirK,其大大低估了环境中nirK的基因丰度。有趣的是,在与本研究相同的采样点利用宏基因组测序技术,对功能微生物的组成与丰度变化特征研究发现,农田土壤中的nirK基因丰度高于nirS基因10倍左右(待发表数据)。
3.2 土壤pH是黑土区氮循环微生物基因丰度空间分布的主要影响因子目前关于当代环境因子与历史进化因素对功能微生物群落地带性分布的相对贡献仍没有统一定论,其检测手段、研究尺度、生态系统类型等因素都会影响研究结果[34]。如Enwall等[35]的研究发现,环境因素是影响反硝化功能基因空间分布异质性的主要原因,而Shi等[36]对区域大尺度下北极土壤功能基因的分布特征的研究发现,北极土壤功能基因的分布主要由空间距离决定并具有尺度依赖性,而土壤环境对功能基因分布特征的贡献较小。
土壤pH是影响土壤微生物群落空间分布格局的最主要驱动因素,被广泛报道[15,37-38]。基于扩增子测序技术,对黑土区农田土壤微生物群落的分布规律研究发现,土壤细菌群落主要受到pH的影响,而真菌群落则主要受到TC含量的制约,说明不同微生物对环境因子的选择性存在差异[15-16]。Wang等[39]研究发现,土壤pH是细菌群落组成变异性最重要的影响因素,但却未对功能基因多样性和组成的空间变异产生显著影响。本研究发现,不同氮循环微生物群落丰度均与土壤pH之间存在显著的正相关性。这与Tsiknia等[40]的研究结果一致,在流域尺度土壤pH与nifH、AOA amoA、AOB amoA、nirK、nirS等基因丰度显著正相关。然而,Avşar和Aras[41]的研究发现,pH与nirK基因丰度存在显著负相关关系,其原因主要是其供试土壤的pH为6.43~8.39,其偏碱性的土壤环境可能了抑制反硝化微生物的生长[42]。此外,基于空间尺度,Chu等[5]对北极微生物的分布格局研究发现,土壤微生物在pH 6.5时,其丰度和多样性最高。本研究VPA和RF分析均显示,土壤pH是氮循环微生物分布的主要影响因素,其主要原因是pH可能直接对土壤微生物施加了一定的生理限制,当pH超出一定范围后将改变微生物间相互作用的结果或者影响那些无法单独生存的微生物类群的生长[43]。其次,土壤pH作为一个综合变量,与一些土壤特性密切相关,如养分可利用性、有机碳特性和土壤水分状况等,它可能并非直接影响微生物的组成,而是通过影响这些土壤条件共同驱动微生物组成的变化[44],这说明除土壤pH外,其他土壤环境因子也是影响微生物群落分布的限制因子。
3.3 氮循环微生物基因丰度存在空间分布格局Bru等[11]研究发现,所有氮循环功能基因没有一种单一的生物地理分布模式,但它们的分布模式存在一个共同的特征,即土壤pH是大多数氮循环功能基因空间分布最重要的驱动因素。尽管土壤pH被证明是微生物地带性分布最主要的制约因素,土壤温度[45]、盐度[46]和土壤养分[47]等因子对不同生态系统微生物的分布格局同样具有制约作用。本研究分析结果显示,除土壤pH外,土壤TC与氮循环微生物功能基因的丰度间也存在显著的正相关关系。Morugán-Coronado等[48]在地中海甜橙果园的研究中证实了氮循环基因的丰度与土壤有机碳呈正相关关系。VPA和RF结果显示,TC对氮循环微生物群落分布起到决定性作用,一些研究也发现TC是影响土壤氮循环微生物群落变异的关键性因子。如Chen等[49]的研究表明,土壤有机碳在决定青藏高原多年冻土区氨氧化微生物和反硝化微生物的丰度和活性方面起着重要作用。同时,这些研究还揭示了土壤水分是驱动土壤功能微生物地理分布的主要因子,因为潮湿的条件有利于有机物的积累[49],进而从侧面印证了土壤碳含量对于功能微生物空间分布的重要性。
本研究中,氮循环微生物功能基因的丰度不仅与土壤pH和TC等土壤环境因子显著相关,也表现出与纬度之间存在正相关关系。VPA分析发现,纬度是除土壤pH和TC外,对氮循环微生物功能群落分异贡献最大的因素。这些结果表明,地理距离对黑土区农田土壤氮循环微生物基因分布有着重要的影响。鉴于黑土区TC含量随纬度升高呈现增加的趋势,说明高纬度地区较高的土壤养分水平使得功能微生物基因具有较高的丰度[13,15]。Wu等[50]研究发现,在大空间尺度下(1 000~4 000 km),地理距离是驱动微生物群落分布的主导因素;而在区域范围内(< 1 000 km),微生物群落分布主要受到环境因子的制约。本研究证实当代环境因子和历史进化因素对功能微生物群落的影响同样具有尺度依赖性,在黑土区740 km的空间范围内,土壤环境因子是驱动氮循环关键过程微生物群落变化的主要因素,同时地理距离也起到了重要的作用。
4 结论基于东北黑土区农田土壤氮循环关键过程微生物基因丰度的空间分布特征的研究结果发现,种植大豆土壤中大豆根系和固氮菌共生体系的存在为根瘤菌提供了独特的生态位,使得种植大豆的非根际土壤nifH基因丰度显著高于种植玉米的土壤样本。反硝化过程nirS、nirK和nosZ基因,固氮nifH基因和氨氧化AOA amoA基因丰度均与土壤pH和TC含量显著正相关。土壤pH对氮循环基因丰度组成存在显著的抑制作用,在中性偏酸的土壤环境中,AOA amoA基因丰度显著高于AOB amoA基因丰度,说明AOA在黑土区氨氧化过程中可能起着主导作用。此外,土壤pH和TC含量作为微生物生长的直接或间接限制因子,不仅影响了东北黑土区氮循环基因组成的分布,而且是氮循环微生物基因空间分布的最主要驱动因子,但地理距离对农田土壤氮循环关键过程微生物基因丰度分布也具有重要影响。
| [1] |
Ravishankara A R, Daniel J S, Portmann R W. Nitrous oxide(N2O): The dominant ozone-depleting substance emitted in the 21st Century[J]. Science, 2009, 326(5949): 123-125. DOI:10.1126/science.1176985
(  0) 0) |
| [2] |
Whitfield J. Biogeography: Is everything everywhere?[J]. Science, 2005, 310(5750): 960-961. DOI:10.1126/science.310.5750.960
(  0) 0) |
| [3] |
Ramette A, Tiedje J M. Multiscale responses of microbial life to spatial distance and environmental heterogeneity in a patchy ecosystem[J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(8): 2761-2766. DOI:10.1073/pnas.0610671104
(  0) 0) |
| [4] |
Zhou J Z, Kang S, Schadt C W, et al. Spatial scaling of functional gene diversity across various microbial taxa[J]. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(22): 7768-7773. DOI:10.1073/pnas.0709016105
(  0) 0) |
| [5] |
Chu H Y, Fierer N, Lauber C L, et al. Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes[J]. Environmental Microbiology, 2010, 12(11): 2998-3006. DOI:10.1111/j.1462-2920.2010.02277.x
(  0) 0) |
| [6] |
King A J, Freeman K R, McCormick K F, et al. Biogeography and habitat modelling of high-alpine bacteria[J]. Nature Communications, 2010, 1: 53. DOI:10.1038/ncomms1055
(  0) 0) |
| [7] |
Griffiths R I, Thomson B C, James P, et al. The bacterial biogeography of British soils[J]. Environmental Microbiology, 2011, 13(6): 1642-1654. DOI:10.1111/j.1462-2920.2011.02480.x
(  0) 0) |
| [8] |
Jiang X L, Liu W Z, Yao L G, et al. The roles of environmental variation and spatial distance in explaining diversity and biogeography of soil denitrifying communities in remote Tibetan wetlands[J]. FEMS Microbiology Ecology, 2020, 96(5): fiaa063. DOI:10.1093/femsec/fiaa063
(  0) 0) |
| [9] |
Hu H W, Zhang L M, Dai Y, et al. pH-dependent distribution of soil ammonia oxidizers across a large geographical scale as revealed by high-throughput pyrosequencing[J]. Journal of Soils and Sediments, 2013, 13: 1439-1449. DOI:10.1007/s11368-013-0726-y
(  0) 0) |
| [10] |
Yao H Y, Campbell C D, Chapman S J, et al. Multi‐factorial drivers of ammonia oxidizer communities: evidence from a national soil survey[J]. Environmental Microbiology, 2013, 15(9): 2545-2556. DOI:10.1111/1462-2920.12141
(  0) 0) |
| [11] |
Bru D, Ramette A, Saby N, et al. Determinants of the distribution of nitrogen-cycling microbial communities at the landscape scale[J]. The ISME Journal, 2011, 5: 532-542. DOI:10.1038/ismej.2010.130
(  0) 0) |
| [12] |
Sun R B, Guo X S, Wang D Z, et al. Effects of long-term application of chemical and organic fertilizers on the abundance of microbial communities involved in the nitrogen cycle[J]. Applied Soil Ecology, 2015, 95: 171-178. DOI:10.1016/j.apsoil.2015.06.010
(  0) 0) |
| [13] |
Zhang X Y, Sui Y Y, Zhang X D, et al. Spatial variability of nutrient properties in black soil of northeast China[J]. Pedosphere, 2007, 17(1): 19-29. DOI:10.1016/S1002-0160(07)60003-4
(  0) 0) |
| [14] |
Liu X M, Li Q, Liang W J, et al. Distribution of soil enzyme activities and microbial biomass along a latitudinal gradient in farmlands of Songliao plain, northeast China[J]. Pedosphere, 2008, 18: 431-440. DOI:10.1016/S1002-0160(08)60034-X
(  0) 0) |
| [15] |
Liu J J, Sui Y Y, Yu Z H, et al. High throughput sequencing analysis of biogeographical distribution of bacterial communities in the black soils of northeast China[J]. Soil Biology and Biochemistry, 2014, 70: 113-122. DOI:10.1016/j.soilbio.2013.12.014
(  0) 0) |
| [16] |
Liu J J, Sui Y Y, Yu Z H, et al. Soil carbon content drives the biogeographical distribution of fungal communities in the black soil zone of northeast China[J]. Soil Biology and Biochemistry, 2015, 83: 29-39. DOI:10.1016/j.soilbio.2015.01.009
(  0) 0) |
| [17] |
Rösch C, Mergel A, Bothe H. Biodiversity of denitrifying and dinitrogen-fixing bacteria in an acid forest soil[J]. Applied and Environmental Microbiology, 2002, 68(8): 3818-3829. DOI:10.1128/AEM.68.8.3818-3829.2002
(  0) 0) |
| [18] |
Francis C A, Roberts K J, Beman M, et al. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean[J]. Proceedings of the National Academy of Sciences of the United States of America, 2005, 102(41): 14683-14688. DOI:10.1073/pnas.0506625102
(  0) 0) |
| [19] |
Rotthauwe J H, Witzel K P, Liesack W. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations[J]. Applied and Environmental Microbiology, 1997, 63(12): 4704-4712. DOI:10.1128/aem.63.12.4704-4712.1997
(  0) 0) |
| [20] |
Throbäck I N, Enwall K, Jarvis Å, et al. Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE[J]. FEMS Microbiology Ecology, 2004, 49(3): 401-417. DOI:10.1016/j.femsec.2004.04.011
(  0) 0) |
| [21] |
Palmer K, Biasi C, Horn M A. Contrasting denitrifier communities relate to contrasting N2O emission patterns from acidic peat soils in arctic tundra[J]. The ISME Journal, 2012, 6: 1058-1077. DOI:10.1038/ismej.2011.172
(  0) 0) |
| [22] |
Rich J J, Heichen R S, Bottomley P J, et al. Community composition and functioning of denitrifying bacteria from adjacent meadow and forest soils[J]. Applied and Environmental Microbiology, 2003, 69(10): 5974-5982. DOI:10.1128/AEM.69.10.5974-5982.2003
(  0) 0) |
| [23] |
Wuchter C, Abbas B, Coolen M J L, et al. Archaeal nitrification in the ocean[J]. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(33): 12317-12322. DOI:10.1073/pnas.0600756103
(  0) 0) |
| [24] |
He J Z, Shen J P, Zhang L M, et al. Quantitative analyses of the abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea of a Chinese upland red soil under long-term fertilization practices[J]. Environmental Microbiology, 2007, 9(9): 2364-2374. DOI:10.1111/j.1462-2920.2007.01358.x
(  0) 0) |
| [25] |
Di H J, Cameron K C, Shen J P, et al. Nitrification driven by bacteria and not archaea in nitrogen-rich grassland soils[J]. Nature Geoscience, 2009, 2: 621-624. DOI:10.1038/ngeo613
(  0) 0) |
| [26] |
Yao H Y, Gao Y M, Nicol G W, et al. Links between ammonia oxidizer community structure, abundance, and nitrification potential in acidic soils[J]. Applied and Environmental Microbiology, 2011, 77(13): 4618-4625. DOI:10.1128/AEM.00136-11
(  0) 0) |
| [27] |
He J Z, Hu H W, Zhang L M. Current insights into the autotrophic thaumarchaeal ammonia oxidation in acidic soils[J]. Soil Biology and Biochemistry, 2012, 55: 146-154. DOI:10.1016/j.soilbio.2012.06.006
(  0) 0) |
| [28] |
Shen X Y, Zhang L M, Shen J P, et al. Nitrogen loading levels affect abundance and composition of soil ammonia oxidizing prokaryotes in semiarid temperate grassland[J]. Journal of Soils and Sediments, 2011, 11: 1243-1252. DOI:10.1007/s11368-011-0375-y
(  0) 0) |
| [29] |
Song H, Che Z, Cao W C, et al. Changing roles of ammonia-oxidizing bacteria and archaea in a continuously acidifying soil caused by over-fertilization with nitrogen[J]. Environmental Science and Pollution Research, 2016, 23: 11964-11974. DOI:10.1007/s11356-016-6396-8
(  0) 0) |
| [30] |
Lee J A, Francis C A. Spatiotemporal characterization of San Francisco Bay denitrifying communities: a comparison of nirK and nirS diversity and abundance[J]. Microbial Ecology, 2017, 73: 271-284. DOI:10.1007/s00248-016-0865-y
(  0) 0) |
| [31] |
Jones C M, Stres B, Rosenquist M, et al. Phylogenetic analysis of nitrite, nitric oxide, and nitrous oxide respiratory enzymes reveal a complex evolutionary history for denitrification[J]. Molecular Biology and Evolution, 2008, 25(9): 1955-1966. DOI:10.1093/molbev/msn146
(  0) 0) |
| [32] |
Bothe H, Ferguson S J, Newton W E. Biology of the nitrogen cycle . Amsterdam: Elsevier Science, 2007: 37-55.
(  0) 0) |
| [33] |
Helen D, Kim H, Tytgat B, et al. Highly diverse nirK genes comprise two major clades that harbour ammonium producing denitrifiers[J]. BMC Genomics, 2016, 17: 155. DOI:10.1186/s12864-016-2465-0
(  0) 0) |
| [34] |
Chu H Y, Wang Y F, Shi Y, et al. Current status and development trend of soil microbial biogeography (In Chinese)[J]. Bulletin of Chinese Academy of Sciences, 2017, 32(6): 585-592. [褚海燕, 王艳芬, 时玉, 等. 土壤微生物生物地理学研究现状与发展态势[J]. 中国科学院院刊, 2017, 32(6): 585-592.]
(  0) 0) |
| [35] |
Enwall K, Throbäck I N, Stenberg M, et al. Soil resources influence spatial patterns of denitrifying communities at scales compatible with land management[J]. Applied and Environmental Microbiology, 2010, 76(7): 2243-2250. DOI:10.1128/AEM.02197-09
(  0) 0) |
| [36] |
Shi Y, Grogan P, Sun H B, et al. Multi-scale variability analysis reveals the importance of spatial distance in shaping Arctic soil microbial functional communities[J]. Soil Biology and Biochemistry, 2015, 86: 126-134. DOI:10.1016/j.soilbio.2015.03.028
(  0) 0) |
| [37] |
Shi Y, Li Y T, Xiang X J, et al. Spatial scale affects the relative role of stochasticity versus determinism in soil bacterial communities in wheat fields across the North China Plain[J]. Microbiome, 2018, 6: 27. DOI:10.1186/s40168-018-0409-4
(  0) 0) |
| [38] |
Feng Y Z, Grogan P, Caporaso J G, et al. pH is a good predictor of the distribution of anoxygenic purple phototrophic bacteria in Arctic soils[J]. Soil Biology and Biochemistry, 2014, 74: 193-200. DOI:10.1016/j.soilbio.2014.03.014
(  0) 0) |
| [39] |
Wang X B, Yao J, Zhang H Y, et al. Environmental and spatial variables determine the taxonomic but not functional structure patterns of microbial communities in alpine grasslands[J]. Science of the Total Environment, 2019, 654: 960-968. DOI:10.1016/j.scitotenv.2018.11.138
(  0) 0) |
| [40] |
Tsiknia M, Paranychianakis N V, Varouchakis E A, et al. Environmental drivers of the distribution of nitrogen functional genes at a watershed scale[J]. FEMS microbiology ecology, 2015, 91(6): fiv052.
(  0) 0) |
| [41] |
Avşar C, Aras E S. Quantification of denitrifier genes population size and its relationship with environmental factors[J]. Archives of Microbiology, 2020, 202(5): 1181-1192. DOI:10.1007/s00203-020-01826-x
(  0) 0) |
| [42] |
Yang Y D, Zhao J, Jiang Y, et al. Response of bacteria harboring nirS and nirK genes to different N fertilization rates in an alkaline northern Chinese soil[J]. European Journal of Soil Biology, 2017, 82: 1-9. DOI:10.1016/j.ejsobi.2017.05.006
(  0) 0) |
| [43] |
Lauber C L, Hamady M, Knight R, et al. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale[J]. Applied and Environmental Microbiology, 2009, 75(15): 5111-5120. DOI:10.1128/AEM.00335-09
(  0) 0) |
| [44] |
Brady N C, Weil R R. The Nature and Properties of Soil . 13.NJ: Prentice Hall, 2007: 960.
(  0) 0) |
| [45] |
Ward D M, Ferris M J, Nold S C, et al. A Natural view of microbial biodiversity within hot spring cyanobacterial mat communities[J]. Microbiology and Molecular Biology Reviews, 1998, 62(4): 1353-1370. DOI:10.1128/MMBR.62.4.1353-1370.1998
(  0) 0) |
| [46] |
Zhao S, Liu J J, Banerjee S, et al. Biogeographical distribution of bacterial communities in saline agricultural soil[J]. Geoderma, 2020, 361: 114095. DOI:10.1016/j.geoderma.2019.114095
(  0) 0) |
| [47] |
Xu Z W, Yu G R, Zhang X Y, et al. Biogeographical patterns of soil microbial community as influenced by soil characteristics and climate across Chinese forest biomes[J]. Applied Soil Ecology, 2018, 124: 298-305. DOI:10.1016/j.apsoil.2017.11.019
(  0) 0) |
| [48] |
Morugán-Coronado A, García-Orenes F, McMillan M, et al. The effect of moisture on soil microbial properties and nitrogen cyclers in Mediterranean sweet orange orchards under organic and inorganic fertilization[J]. Science of the Total Environment, 2019, 655: 158-167. DOI:10.1016/j.scitotenv.2018.11.174
(  0) 0) |
| [49] |
Chen Y L, Kou D, Li F, et al. Linkage of plant and abiotic properties to the abundance and activity of N-cycling microbial communities in Tibetan permafrost-affected regions[J]. Plant Soil, 2019, 434: 453-466.
(  0) 0) |
| [50] |
Wu B, Tian J Q, Bai C M, et al. The biogeography of fungal communities in wetland sediments along the Changjiang River and other sites in China[J]. The ISME Journal, 2013, 7: 1299-1309.
(  0) 0) |
 2022, Vol. 59
2022, Vol. 59


