2. 农业农村部长江下游平原农业环境重点实验室, 江苏省农业科学院农业资源与环境研究所, 南京 210014;
3. 中国科学院大学, 北京 100049;
4. Department of Plant, Soil and Microbial Sciences, Michigan State University, MI 48824, USA
2. Key Laboratory of Agro-Environment in Downstream of Yangtze Plain, Ministry of Agriculture and Rural Affairs, Institute of Agricultural Resources and Environment, Jiangsu Academy of Agricultural Sciences, Nanjing 210014, China;
3. University of the Chinese Academy of Sciences, Beijing 100049, China;
4. Department of Plant, Soil and Microbial Sciences, Michigan State University, MI 48824, USA
抗生素为保证人类健康、动物生长和提高农业生产做出了巨大贡献。然而,随着人类活动和畜禽养殖过程中长期过度使用和滥用抗生素,抗生素抗性基因(Antibiotic resistance genes,ARGs)在全世界范围内广泛传播,抗生素抗性基因已被认为是人类在本世纪面临的六大新环境问题和全球性挑战问题之首[1-3]。即使低浓度下的抗生素残留也会对细菌产生选择性压力,伴随着人类活动的加剧,抗生素抗性细菌(Antibiotic-resistant bacteria,ARB)和抗生素抗性基因在环境中广泛共存(包括畜禽养殖排泄物、废水、污泥、大气和土壤等),前者具有普遍性和多样性,后者具有可转移性[4]。ARGs可以通过垂直转移和水平基因转移(Horizontal gene transfer,HGT)在亲本和子代之间或不同种类的细菌之间传播,导致其在环境介质进行转移,其中水平基因转移被认为是ARGs传播的主要途径[5]。
抗生素抗性在环境中的传播与ARB和ARGs极为相关,其中,土壤中DNA序列丰度最高、微生物种类最多,微生物在交流和竞争过程中,通过代谢活动不断产生抗生素、毒素等代谢产物,当前临床医疗和畜禽养殖过程中使用的大部分抗生素均来源于土壤中的微生物,其对抗生素抗性的产生和传播具有重要的影响。此外,土壤中的重金属等污染物对ARGs的协同选择作用以及移动基因元件(Mobile genetic elements,MGEs)均能够促进土壤中ARGs的传播。携带ARGs的微生物死亡后,体内的DNA释放到环境中,在土壤矿物和有机胶体的保护下,能够长期存在于土壤环境中,容易被其他微生物吸收而获得抗生素抗性[6]。
环境中抗生素抗性研究需要遵循“大健康(One Health)”准则,包括跨学科的认知和三个主要领域的综合研究,即人类健康、动物健康和环境健康[3-7]。其中,土壤是“大健康”准则中的重要组成部分,其自身具有多种天然抗性,同时由于人类的农业生产和临床医疗等活动,造成大量抗性基因汇入土壤,土壤已然成为环境中ARB和ARGs的储存库。此外,ARB和ARGs还可以通过多种途径转移到人类和动、植物体内[8](图 1),从而引发一系列土壤生态风险和人体健康问题。
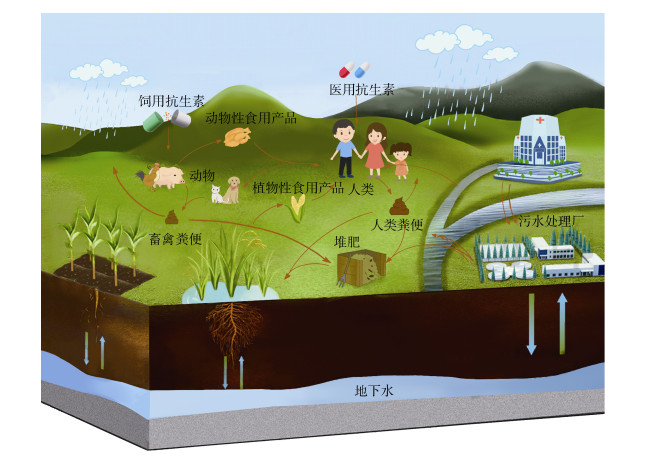
|
图 1 抗生素抗性细菌和抗生素抗性基因在环境中的传播 Fig. 1 The spread of antibiotic-resistant bacteria and antibiotic resistance genes in the environment |
微生物的“内在抗性”是指存在于微生物基因组上的抗性基因或尚未表达抗性的抗性基因[9],微生物在一定环境中还可通过随机突变而获得抗生素抗性,微生物“内在抗性”是环境中ARGs的重要组成部分[10]。土壤环境中存在大量由微生物合成的天然抗生素,形成一定的环境压力,使微生物种群获得内在抗性,其中微生物的内在抗性是未受人类活动影响土壤环境中ARGs的主要来源。
20世纪初,在抗生素时代被提出之前,ARGs就已经在环境中存在和传播[11]。在远离人类活动影响的阿拉斯加冻结土壤中,多种新β-内酰胺酶和氯霉素类抗性基因被发现,并首次检测到双功能β-内酰胺酶抗性基因,表明该抗性基因能够自然存在于环境中[12]。在3万年前的白令永久冻土中,晚更新世生物体内DNA中存在着高度多样性的ARGs,对β-内酰胺类、四环素类和糖肽类抗生素具有抗药性[11]。在南极洲的冈瓦纳大陆研究站和新建Jang Bogo研究站共检测到8个主要ARGs类别,包括73个ARGs和MGEs(整合子、转座子、质粒和基因盒)[13]。以上研究均证实,在抗生素被人类广泛使用之前,ARGs就已经存在于土壤环境中。随着第二代高通量测序技术和第三代单分子实时测序和纳米孔测序等测序技术的发展,人们能够更加全面且准确地揭示不同环境中ARGs以及与ARGs水平转移相关的基因元件的分布水平。
1.2 外部来源由于人类活动的加剧,来自人类和动物的新型致病菌和抗生素进入土壤环境,成为重要的环境污染物[14]。从而促使ARGs在环境中广泛传播,其中与人类活动相关的ARGs在土壤环境中逐渐富集[15-16]。
随着畜牧业一体化的发展,规模化畜禽养殖场是动物粪便的主要来源。畜禽养殖场动物肠道和粪便中含有丰富的ARB和ARGs,粪便堆肥施用可能是动物体内ARGs进入土壤环境的主要途径[17-18]。粪肥及其堆肥中ARGs的种类呈现多样性,对四环素类、β-内酰胺类、氨基糖苷类、氯霉素、磺胺类抗生素等多种药物具有抗性[19-20]。堆肥施用过程中,粪肥中许多ARGs均由质粒或整合子携带,ARGs与MGEs共存能够促进ARGs的转移和传播[21]。粪肥中携带ARGs的外来微生物进入土壤往往难以与土著微生物竞争存活,其释放到环境中的ARGs有可能通过水平基因转移进入土著微生物体内而使其获得抗生素抗性。粪肥中的有机质和养分能够促进微生物生长,导致ARGs在土壤环境中持续富集[22]。此外,粪肥携带的抗生素残留对土壤微生物造成了持续性的选择压力,导致微生物体内ARGs被诱导表达,同时固有基因也有可能会发生基因突变产生新的ARGs,促使共生微生物和病原微生物之间的基因交换,从而增加ARGs和MGEs从粪肥向土壤环境传播的风险[19-23]。
废水的再利用,是解决水资源短缺问题的一种实用的解决方案,可显著缓解水资源压力。尤其是在农业生产中,污水经污水处理厂处理后直接排入河流或用于农业灌溉。然而,医疗废水和动物养殖场污水中的ARGs进入污水处理系统后,现有水处理技术不能彻底去除抗生素及ARGs,污水处理厂的出水中仍然富含抗生素、病原微生物和营养物质,被视作ARGs选择和转移的高风险“热点”区域[24],同时也是其他水体环境中ARGs最重要的来源之一。在我国南部珠江三角洲地区,采用鱼塘水灌溉土壤中残留的四环素和磺胺甲嘧啶及其相应的ARGs显著高于生活污水。灌溉水中抗生素及其ARGs的浓度显著高于灌溉土壤,这表明废水是土壤环境中抗生素及ARGs的主要来源[25]。使用未经处理的污水作为灌溉水源,土壤中ARGs的绝对丰度普遍提高两个数量级(除tetQ、aadA、intI1、qacE+qacE△I和IncP-1质粒)[26]。在澳大利亚维多利亚州地区,经处理后废水灌溉的城市公园土壤中共鉴定出40个特有的ARGs,其中β-内酰胺类抗性基因最为普遍[27]。
在污水处理过程中,污泥能够大量富集抗生素和ARGs。活性污泥中ARGs的含量相较于进水样品增加了947倍,这可能是由于微生物量的增加导致的结果[28]。因此,污泥是微生物ARGs和MGEs富集的“热点”区域。堆肥还田是污泥处置和再生产品使用的重要方式,在此过程中携带移动基因元件的外来微生物与土著微生物通过水平基因转移共享遗传信息,也是ARGs向农田土壤中扩散的关键途径[29]。相比于未改良土壤,经污泥改良后的土壤中,尽管ARGs的相对丰度无明显变化,但绝对丰度在20 d内有短期增加[30]。
2 土壤中抗性基因的传播和影响因素 2.1 土壤中抗性基因的传播土壤中富集的ARGs能够通过多途径传播到水、大气等介质,厘清ARGs在不同介质中的传播规律及其影响因素对于有效评估环境中ARGs的生态风险尤为重要。
在淋溶和渗滤作用下,土壤中的ARB和ARGs能够转移至地表水、地下水和饮用水[31],导致水环境中存在多种与畜禽养殖等人类活动密切相关的ARGs,严重威胁人体健康。目前,水体环境及水生生物体内的ARGs污染已经十分普遍。不仅在饮用水中检测到四环素类和磺胺类抗性基因,在饮用水源中分离的Enterobacteriaceae菌株中也有4种ARGs被检测出[32-33]。Koike等[34]发现,养猪场附近的池塘和地下水中同时存在7种四环素抗性基因(tetM、tetO、tetQ、tetW、tetC、tetH和tetZ),其中地下水中的基因序列与池塘中的基因序列高度一致,甚至在地下水中还发现了新的序列簇和独特的抗性基因库。
土壤中的ARGs能够以生物气溶胶的形式进入到大气中,在风力、气流等作用下,空气中的ARGs在不同的地区远距离传播,之后通过干/湿沉降传播至受人类影响较小的偏远地区土壤,从而导致ARGs在全球范围传播[35]。污染严重的空气中颗粒物浓度较高,为ARB和ARGs提供了更多的附着点[36]。例如,在畜禽养殖及其粪便处理过程中,土壤中大量的污染物和ARB极易发生雾化,导致地球表层成为ARGs重要的载体和储存库,从而促使ARGs在土壤与大气之间发生传播[37]。
土壤微生物组是植物获得ARGs的重要来源,在根际环境中,水通量增加和养分输入可以刺激微生物代谢以及体内质粒的转移接合,促进抗性细菌从土壤微生物组向植物微生物组迁移,根际和叶际环境成为植物界微生物基因转移的“热点”区域[38-39]。众多研究发现,植物内生菌(根际内生菌和叶际内生菌)能够通过多种途径从施加粪肥的土壤中获得抗生素抗性,是抗性基因组和移动基因元件的主要载体和传播者[40]。经堆肥处理后,土壤中的ARGs具有转移到蔬菜体内的风险,尤其需要注意的是生食或轻加工的蔬菜[41]。例如,猪粪堆肥施用增加了胡萝卜组织尤其是外表皮中ARGs的累积,食用前去皮是降低人体健康风险的有效策略[42]。此外,植物界中潜在的可转移基因库与宿主密切相关,具有高度的流通性[39]。由于这些“热点”区域处于非均质的动态环境,如根际微生物及其分泌物(糖、有机酸和氨基酸等)可能会对ARGs从土壤向植物体内的迁移产生影响[43],在不同条件下难以评估这些因素对ARGs水平基因转移的影响。
2.2 土壤中抗性基因传播的影响因素临床医疗和畜禽养殖活动中抗生素的大量使用,以及农业生产中动物粪便和污泥的处理再利用,对ARGs传播产生了关键性的选择压力。此外,其他环境自然因素也影响微生物的抗生素抗性,如营养匮乏、极端温度胁迫和氧化还原条件等[44-45]。其中,土壤理化性质、农艺调控及环境污染物等对土壤中ARGs的传播起到至关重要的作用。
土壤理化性质(pH、温度、有机质、含水率等)可以显著影响土壤中微生物群落的结构多样性和功能,已被确定为诱导抗生素抗性进化和传播的重要环境因素[46]。在长期施有机肥土壤中,ARGs的丰度与抗生素浓度和土壤理化性质(pH和有机质)紧密相关,且随着土壤深度的增加,四环素类药物浓度和ARGs丰度普遍降低[47]。Wu等[48]报道,在猪饲养场附近的土壤中,tet基因绝对拷贝数不仅与土壤中四环素残留浓度显著相关,还受土壤有机质等环境因子的影响。土壤pH通过影响养分有效性或微生物生理活性对其产生了较大的选择性压力,并对微生物的丰度和多样性产生影响,进而影响了ARGs在土壤环境中的传播[49]。土壤含水率也能够通过影响微生物活性来干扰抗生素和ARGs的去除,同时含水率改变也会影响土壤间隙度、吸附系数和离子电位,从而影响土壤中ARGs的持久性[50]。
常见的农艺调控措施,如施肥、种植模式、灌溉水源和农药喷施等会影响土壤固有的抗生素抗性。不同的施肥和种植体系(如旱地和水稻中)能够影响土壤中ARGs和MGEs的丰度水平。有机粪肥中含有适应细菌生长的丰富碳源,并且含有抗生素、重金属、有机污染物等共选择物质,能够促进抗生素抗性在土壤环境中的传播[51]。尤其是农业土壤在长期施用堆肥的过程中,土壤中的ARGs水平在初始阶段呈上升趋势,显著增加了粪肥中ARGs向土壤环境中传播的风险[52]。Wang等[53]报道,在水稻和旱地两种种植体系中,水稻土中的ARGs积累量和相应的微生物量高于旱地土壤,其中堆肥施用对两种种植体系土壤中的ARGs累积效果均不显著,而化肥施用增加了旱地土壤中ARGs的相对丰度,降低了水稻土中ARGs的相对丰度。此外,草甘膦、草铵膦和草甘菊等除草剂的喷施同样能够改变土壤微生物群落的遗传组成,促使ARGs和MGEs土壤中累积。除草剂暴露还提高了多药抗性质粒的细胞膜通透性和偶联频率,促进了ARGs在微生物间的交换[54]。与此同时,生物炭作为土壤改良剂已被广泛应用于农业生产过程,其中抗生素能够和生物炭上的芳香环之间通过π-π键发挥相互作用,从而驱动土壤微生物间ARGs的选择或共选择[55]。生物炭改良剂普遍被认为可显著降低非种植土壤中ARGs的丰度,其潜在机制可归因于生物炭的吸附作用降低抗生素和重金属的流动性,同时生物炭的添加还能够影响细菌群落结构的变化和抗体的产生[56-58]。
土壤中的污染物(重金属、微塑料、纳米颗粒等)能够显著影响ARGs的类型、丰度和迁移过程,导致ARGs在污染土壤中传播风险的不可预测性。重金属可通过共抗性、交叉抗性、共调控作用对环境中ARGs产生选择性压力,从而使土壤中ARGs丰度维持较高水平[59]。与抗生素不同,重金属在环境中长期存在,了解重金属对抗生素抗性和对环境中细菌群落结构变化的驱动至关重要[60]。微塑料比表面积大、吸附性强,可吸附环境中的重金属、抗生素、抗性基因和抗性细菌等污染物,微塑料逐渐成为微生物抗生素抗性的新“热点”区域[61-62]。例如,在设施蔬菜土壤中,微塑料的粒径越大、风化作用越强或蔬菜种植期越长,其能够从周围环境中吸附更多的抗生素、重金属和移动基因元件,从而提高微塑料表面ARGs的种类和数量[63]。研究表明微塑料对ARGs的富集量与微塑料的浓度成反比,能够大量富集环境中的ARGs,特别是胞外ARGs,从而显著提高ARGs的迁移能力[64]。尽管纳米颗粒广泛存在于土壤、废水和污泥等多种环境中,但其对ARB和ARGs的影响还缺乏全面了解。Zhu等[65]发现AgNPs暴露能够干扰跳虫肠道细菌群落的组成,降低弹尾虫肠道微生物群中ARGs的发生率。然而,Qiu等[66]报道纳米Al2O3的使用显著促进了质粒RP4、RK2和pCF10携带的耐药基因的水平基因转移,这可能是由于纳米Al2O3能够通过诱导氧化应激损伤细菌细胞膜,从而增强接合基因表达。该结果表明纳米材料可能会促进敏感细菌获得抗生素抗性,从而带来一定的环境和人体健康风险。
3 土壤中抗性基因的消减技术为了减少土壤、水、大气等环境介质中ARGs的传播,降低环境中抗生素抗性的风险,科研人员重点研发各种ARGs消减技术,如好氧堆肥、厌氧发酵和水处理工艺,其目的是减少有机肥、污泥还田、污水灌溉等农业生产过程造成的土壤中抗生素和外源性ARGs的输入和富集。其中,环境中ARGs的削减主要与细胞的运输、宿主的死亡以及胞外ARGs的衰减有关[46,67]。
好氧堆肥技术常被用于畜禽粪便和污泥的无害化处理以及肥料的资源化利用,同时也能够有效削减畜禽粪便中的ARGs,其中,温度等处理工艺影响着有机废弃物中ARGs的丰度。连续高温堆肥过程中,温度的变化能够显著降低动物粪便中ARGs和整合子的多样性和丰度[68]。此外,沸石、过磷酸钙和硫酸亚铁的添加能够有效去除畜禽粪便中的ARGs,去除率分别达到86.5%、68.6%和72.2%[69],此外,生物炭和鼠李糖脂、吐温80等表面活性剂的添加也能够对ARGs的去除起到一定作用[70-71]。然而,好氧堆肥技术难以去除所有类型的ARGs,好氧堆肥产品仍然是ARGs的重要储存库[22]。
厌氧发酵技术也常被用于处理畜禽粪便和污泥等有机固体废弃物。畜禽粪便经厌氧发酵后,可以减少动物粪便和有机污泥中ARB和ARGs的数量和丰度,并且其残渣可作为有机肥施用,是农业废弃物中ARGs去除和实现可持续性利用的有效方法[72]。Sun等[73]通过采用预处理、高温消化、两段消化、添加剂和固态消化等处理手段,研究厌氧发酵对ARGs的去除效果。其中,操作参数对ARGs的去除效率起着至关重要的作用,例如在污泥厌氧消化过程中,温度越高、水力停留时间越长,ARGs去除率越高[74]。
水处理工艺技术亦能够有效削减污水、污泥以及饮用水中的ARGs。在废水处理过程中,紫外线杀菌、氯气和臭氧消毒等技术被用来诱导细菌损伤,从而有效去除水体环境中的ARGs[75]。抗性细菌的杀菌过程与消毒剂的相对活性和重要细胞成分(包括氨基酸、糖类、脂类和核酸)的消耗量密切相关[76]。在不同的紫外线或氯离子浓度下测定废水中ARGs的转移频率,结果表明紫外线和氯化消毒能够影响ARGs的接合转移,其中紫外线强度和氯气剂量是去除ARGs的重要因素[77]。已有研究发现,污水处理厂各处理环节对ARGs去除的影响不同,例如,生物处理和化学处理能够造成ARGs丰度发生不同变化[78]。在废水处理过程中,尽管抗性细菌因细胞结构遭到破坏而失活,但ARGs仍可能存在于细胞碎片中,对人体健康构成潜在的风险。因此,需要严格评估现有的水处理工艺,以发挥它们在缓解环境中ARGs风险的潜力。
4 土壤中抗性基因与人体健康大部分与临床相关的ARGs来自于环境中的微生物[79],同时土壤微生物携带的ARGs也存在于人类临床病原体,以及对磺胺类、氨基糖苷类和β-内酰胺类等药物具有高水平抗性的新型细菌体内[80]。采用宏基因组技术对土壤微生物进行功能分析,研究发现土壤微生物和人类临床病原体的耐药组核苷酸同源性较高(> 99%),由此推断不同环境中微生物之间的ARGs可能存在水平基因转移。在自然和受人类影响的环境中,整合子被证实是多种ARGs的主要载体[14]。例如,intI1属于Ⅰ类整合子,与同一基因位点上多个ARGs的整合密切相关,促进了ARGs的水平基因转移过程[21]。
目前,关于ARGs从环境微生物向人类病原体转移的认识仍存在一定局限性。土壤中施加有机肥后,在叶菜(芝麻菜、香菜)表面检测到多种ARGs,该ARGs极易通过摄食过程进入人类体内,表明土壤环境、植物和人类之间存在直接的食物链联系[81-82]。将生菜或香菜叶富集培养,经检测在人体大肠杆菌中发现多个抗性质粒[81]。当细菌基因组中的多个ARGs同时出现在质粒上时,“超级细菌”可能会在全球范围内爆发。例如,黏菌素曾用作应对多重耐药细菌爆发威胁的最后一道防线,但如今,我国上海首次报道在动物和人类肠道内发现携带黏菌素抗性基因mcr-1的质粒,随后该抗性基因出现在世界各地人类病原体、家禽、猪肉等样本中,导致其在全球范围内广泛传播[83]。
5 结论与展望目前,由于人类活动和畜禽养殖过程中长期过度使用和滥用抗生素,土壤中ARB和ARGs污染已经广泛存在,但其生态风险缺乏严格管控,难以确定土壤环境中ARB和ARGs的最大允许阈值,更难以客观地评估与其相关的人类健康风险。同时,关于土壤中ARGs的传播阻控和削减技术研究仍然不足。因此,在“大健康”准则下,亟需建立一个公开的全球网络监测预警系统,能够持续掌握抗生素生产使用情况以及评估农业生产和临床医疗活动中抗生素抗性的潜在风险;进一步厘清ARGs在土壤环境中的传播和扩散机制;在此基础上研发新型高效的ARGs传播阻断和消减技术,从而控制土壤中抗生素抗性的产生和传播。
| [1] |
Fair R J, Tor Y. Antibiotics and bacterial resistance in the 21st century[J]. Perspectives in Medicinal Chemistry, 2014, 6: 25-64.
(  0) 0) |
| [2] |
Pruden A, Pei R T, Storteboom H, et al. Antibiotic resistance genes as emerging contaminants: Studies in northern Colorado[J]. Environmental Science & Technology, 2006, 40(23): 7445-7450.
(  0) 0) |
| [3] |
Tiedje J M, Wang F, Manaia C M, et al. Antibiotic resistance genes in the human-impacted environment: A One Health perspective[J]. Pedosphere, 2019, 29(3): 273-282. DOI:10.1016/S1002-0160(18)60062-1
(  0) 0) |
| [4] |
Martínez J L. Antibiotics and antibiotic resistance genes in natural environments[J]. Science, 2008, 321(5887): 365-367. DOI:10.1126/science.1159483
(  0) 0) |
| [5] |
Lerminiaux N A, Cameron A D S. Horizontal transfer of antibiotic resistance genes in clinical environments[J]. Canadian Journal of Microbiology, 2019, 65(1): 34-44. DOI:10.1139/cjm-2018-0275
(  0) 0) |
| [6] |
Hu X J, Sheng X, Zhang W, et al. Nonmonotonic effect of montmorillonites on the horizontal transfer of antibiotic resistance genes to bacteria[J]. Environmental Science & Technology Letters, 2020, 7(6): 421-427.
(  0) 0) |
| [7] |
Wang F, Fu Y H, Sheng H J, et al. Antibiotic resistance in the soil ecosystem: A One Health perspective[J]. Current Opinion in Environmental Science & Health, 2021, 20: 100230.
(  0) 0) |
| [8] |
Zhu Y G, Zhao Y, Zhu D, et al. Soil biota, antimicrobial resistance and planetary health[J]. Environment International, 2019, 131: 105059. DOI:10.1016/j.envint.2019.105059
(  0) 0) |
| [9] |
Davies J, Davies D. Origins and evolution of antibiotic resistance[J]. Microbiology and Molecular Biology Reviews, 2010, 74(3): 417-433. DOI:10.1128/MMBR.00016-10
(  0) 0) |
| [10] |
Su J Q, Huang F Y, Zhu Y G. Antibiotic resistance genes in the environment (In Chinese)[J]. Biodiversity Science, 2013, 21(4): 481-487. [苏建强, 黄福义, 朱永官. 环境抗生素抗性基因研究进展[J]. 生物多样性, 2013, 21(4): 481-487.]
(  0) 0) |
| [11] |
D'Costa V M, King C E, Kalan L, et al. Antibiotic resistance is ancient[J]. Nature, 2011, 477(7365): 457-461. DOI:10.1038/nature10388
(  0) 0) |
| [12] |
Allen H K, Moe L A, Rodbumrer J, et al. Functional metagenomics reveals diverse β-lactamases in a remote Alaskan soil[J]. The ISME Journal, 2009, 3(2): 243-251. DOI:10.1038/ismej.2008.86
(  0) 0) |
| [13] |
Wang F, Stedtfeld R D, Kim O S, et al. Influence of soil characteristics and proximity to Antarctic research stations on abundance of antibiotic resistance genes in soils[J]. Environmental Science & Technology, 2016, 50(23): 12621-12629.
(  0) 0) |
| [14] |
Forsberg K J, Reyes A, Wang B, et al. The shared antibiotic resistome of soil bacteria and human pathogens[J]. Science, 2012, 337(6098): 1107-1111. DOI:10.1126/science.1220761
(  0) 0) |
| [15] |
Bengtsson-Palme J, Kristiansson E, Larsson D G J. Environmental factors influencing the development and spread of antibiotic resistance[J]. FEMS Microbiology Reviews, 2017, 42(1): fux053.
(  0) 0) |
| [16] |
Peterson E, Kaur P. Antibiotic resistance mechanisms in bacteria: Relationships between resistance determinants of antibiotic producers, environmental bacteria, and clinical pathogens[J]. Frontiers in Microbiology, 2018, 9: 2928. DOI:10.3389/fmicb.2018.02928
(  0) 0) |
| [17] |
Zheng F, Bi Q F, Giles M, et al. Fates of antibiotic resistance genes in the gut microbiome from different soil fauna under long-term fertilization[J]. Environmental Science & Technology, 2021, 55(1): 423-432.
(  0) 0) |
| [18] |
Zhu D, Chen Q L, Ding J, et al. Antibiotic resistance genes in the soil ecosystem and planetary health: Progress and prospect (In Chinese)[J]. Scientia Sinica: Vitae, 2019, 49(12): 1652-1663. [朱冬, 陈青林, 丁晶, 等. 土壤生态系统中抗生素抗性基因与星球健康: 进展与展望[J]. 中国科学: 生命科学, 2019, 49(12): 1652-1663.]
(  0) 0) |
| [19] |
Chen C Q, Pankow C A, Oh M, et al. Effect of antibiotic use and composting on antibiotic resistance gene abundance and resistome risks of soils receiving manure- derived amendments[J]. Environment International, 2019, 128: 233-243. DOI:10.1016/j.envint.2019.04.043
(  0) 0) |
| [20] |
Xie W Y, Yang X P, Li Q, et al. Changes in antibiotic concentrations and antibiotic resistome during commercial composting of animal manures[J]. Environmental Pollution, 2016, 219: 182-190. DOI:10.1016/j.envpol.2016.10.044
(  0) 0) |
| [21] |
Ma L P, Li A D, Yin X L, et al. The prevalence of integrons as the carrier of antibiotic resistance genes in natural and man-made environments[J]. Environmental Science & Technology, 2017, 51(10): 5721-5728.
(  0) 0) |
| [22] |
Qian X, Sun W, Gu J, et al. Variable effects of oxytetracycline on antibiotic resistance gene abundance and the bacterial community during aerobic composting of cow manure[J]. Journal of Hazardous Materials, 2016, 315: 61-69. DOI:10.1016/j.jhazmat.2016.05.002
(  0) 0) |
| [23] |
You Y Q, Silbergeld E K. Learning from agriculture: Understanding low-dose antimicrobials as drivers of resistome expansion[J]. Frontiers in Microbiology, 2014, 5: 284.
(  0) 0) |
| [24] |
Gatica J, Cytryn E. Impact of treated wastewater irrigation on antibiotic resistance in the soil microbiome[J]. Environmental Science and Pollution Research International, 2013, 20(6): 3529-3538. DOI:10.1007/s11356-013-1505-4
(  0) 0) |
| [25] |
Pan M, Chu L M. Occurrence of antibiotics and antibiotic resistance genes in soils from wastewater irrigation areas in the Pearl River Delta region, Southern China[J]. Science of the Total Environment, 2018, 624: 145-152. DOI:10.1016/j.scitotenv.2017.12.008
(  0) 0) |
| [26] |
Jechalke S, Broszat M, Lang F, et al. Effects of 100 years wastewater irrigation on resistance genes, class 1 integrons and IncP-1 plasmids in Mexican soil[J]. Frontiers in Microbiology, 2015, 6: 163.
(  0) 0) |
| [27] |
Han X M, Hu H W, Shi X Z, et al. Impacts of reclaimed water irrigation on soil antibiotic resistome in urban parks of Victoria, Australia[J]. Environmental Pollution, 2016, 211: 48-57. DOI:10.1016/j.envpol.2015.12.033
(  0) 0) |
| [28] |
An X L, Su J Q, Li B, et al. Tracking antibiotic resistome during wastewater treatment using high throughput quantitative PCR[J]. Environment International, 2018, 117: 146-153. DOI:10.1016/j.envint.2018.05.011
(  0) 0) |
| [29] |
Chen H, Zhang M M. Occurrence and removal of antibiotic resistance genes in municipal wastewater and rural domestic sewage treatment systems in Eastern China[J]. Environment International, 2013, 55: 9-14. DOI:10.1016/j.envint.2013.01.019
(  0) 0) |
| [30] |
Zhou Z, Raskin L, Zilles J L. Effects of Swine manure on macrolide, lincosamide, and streptogramin B antimicrobial resistance in soils[J]. Applied and Environmental Microbiology, 2010, 76(7): 2218-2224. DOI:10.1128/AEM.02183-09
(  0) 0) |
| [31] |
Chee-Sanford J C, Mackie R I, Koike S, et al. Fate and transport of antibiotic residues and antibiotic resistance genes following land application of manure waste[J]. Journal of Environmental Quality, 2009, 38(3): 1086-1108. DOI:10.2134/jeq2008.0128
(  0) 0) |
| [32] |
Rizzo L, Manaia C, Merlin C, et al. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review[J]. Science of the Total Environment, 2013, 447: 345-360. DOI:10.1016/j.scitotenv.2013.01.032
(  0) 0) |
| [33] |
Tao R, Ying G G, Su H C, et al. Detection of antibiotic resistance and tetracycline resistance genes in Enterobacteriaceae isolated from the Pearl Rivers in South China[J]. Environmental Pollution, 2010, 158(6): 2101-2109. DOI:10.1016/j.envpol.2010.03.004
(  0) 0) |
| [34] |
Koike S, Krapac I G, Oliver H D, et al. Monitoring and source tracking of tetracycline resistance genes in lagoons and groundwater adjacent to swine production facilities over a 3-year period[J]. Applied and Environmental Microbiology, 2007, 73(15): 4813-4823. DOI:10.1128/AEM.00665-07
(  0) 0) |
| [35] |
Zhu G, Wang X, Yang T, et al. Air pollution could drive global dissemination of antibiotic resistance genes[J]. The ISME Journal, 2021, 15(1): 270-281. DOI:10.1038/s41396-020-00780-2
(  0) 0) |
| [36] |
Gao M, Jia R Z, Qiu T L, et al. Size-related bacterial diversity and tetracycline resistance gene abundance in the air of concentrated poultry feeding operations[J]. Environmental Pollution, 2017, 220: 1342-1348. DOI:10.1016/j.envpol.2016.10.101
(  0) 0) |
| [37] |
Dungan R S. Fate and transport of bioaerosols associated with livestock operations and manures[J]. Journal of Animal Science, 2010, 88(11): 3693-3706. DOI:10.2527/jas.2010-3094
(  0) 0) |
| [38] |
Kroer N, Barkay T, Sørensen S, et al. Effect of root exudates and bacterial metabolic activity on conjugal gene transfer in the rhizosphere of a marsh plant[J]. FEMS Microbiology Ecology, 1998, 25(4): 375-384. DOI:10.1111/j.1574-6941.1998.tb00489.x
(  0) 0) |
| [39] |
van Elsas J D, Turner S, Bailey M J. Horizontal gene transfer in the phytosphere[J]. New Phytologist, 2003, 157(3): 525-537. DOI:10.1046/j.1469-8137.2003.00697.x
(  0) 0) |
| [40] |
Zhang H, Li X N, Yang Q X, et al. Plant growth, antibiotic uptake, and prevalence of antibiotic resistance in an endophytic system of pakchoi under antibiotic exposure[J]. International Journal of Environmental Research and Public Health, 2017, 14(11): 1336. DOI:10.3390/ijerph14111336
(  0) 0) |
| [41] |
Marti R, Scott A, Tien Y C, et al. Impact of manure fertilization on the abundance of antibiotic-resistant bacteria and frequency of detection of antibiotic resistance genes in soil and on vegetables at harvest[J]. Applied and Environmental Microbiology, 2013, 79(18): 5701-5709. DOI:10.1128/AEM.01682-13
(  0) 0) |
| [42] |
Mei Z, Xiang L L, Wang F, et al. Bioaccumulation of manure-borne antibiotic resistance genes in carrot and its exposure assessment[J]. Environment International, 2021, 157: 106830. DOI:10.1016/j.envint.2021.106830
(  0) 0) |
| [43] |
Berendsen R L, Pieterse C M J, Bakker P A H M. The rhizosphere microbiome and plant health[J]. Trends in Plant Science, 2012, 17(8): 478-486. DOI:10.1016/j.tplants.2012.04.001
(  0) 0) |
| [44] |
Cruz-Loya M, Kang T M, Lozano N A, et al. Stressor interaction networks suggest antibiotic resistance co-opted from stress responses to temperature[J]. The ISME Journal, 2019, 13(1): 12-23. DOI:10.1038/s41396-018-0241-7
(  0) 0) |
| [45] |
Poole K. Stress responses as determinants of antimicrobial resistance in Gram-negative bacteria[J]. Trends in Microbiology, 2012, 20(5): 227-234. DOI:10.1016/j.tim.2012.02.004
(  0) 0) |
| [46] |
Chen Z Y, Zhang W, Wang G, et al. Bioavailability of soil-sorbed tetracycline to Escherichia coli under unsaturated conditions[J]. Environmental Science & Technology, 2017, 51(11): 6165-6173.
(  0) 0) |
| [47] |
Tang X J, Lou C L, Wang S X, et al. Effects of long-term manure applications on the occurrence of antibiotics and antibiotic resistance genes(ARGs)in paddy soils: Evidence from four field experiments in south of China[J]. Soil Biology & Biochemistry, 2015, 90: 179-187.
(  0) 0) |
| [48] |
Wu N, Qiao M, Zhang B, et al. Abundance and diversity of tetracycline resistance genes in soils adjacent to representative swine feedlots in China[J]. Environmental Science & Technology, 2010, 44(18): 6933-6939.
(  0) 0) |
| [49] |
Xiao K Q, Li B, Ma L P, et al. Metagenomic profiles of antibiotic resistance genes in paddy soils from South China[J]. FEMS Microbiology Ecology, 2016, 92(3): fiw023. DOI:10.1093/femsec/fiw023
(  0) 0) |
| [50] |
Ingerslev F, Toräng L, Loke M L, et al. Primary biodegradation of veterinary antibiotics in aerobic and anaerobic surface water simulation systems[J]. Chemosphere, 2001, 44(4): 865-872. DOI:10.1016/S0045-6535(00)00479-3
(  0) 0) |
| [51] |
Zhu Y G, Johnson T A, Su J Q, et al. Diverse and abundant antibiotic resistance genes in Chinese swine farms[J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(9): 3435-3440. DOI:10.1073/pnas.1222743110
(  0) 0) |
| [52] |
Xu M, Stedtfeld R D, Wang F, et al. Composting increased persistence of manure-borne antibiotic resistance genes in soils with different fertilization history[J]. Science of the Total Environment, 2019, 689: 1172-1180. DOI:10.1016/j.scitotenv.2019.06.376
(  0) 0) |
| [53] |
Wang F, Xu M, Stedtfeld R D, et al. Long-term effect of different fertilization and cropping systems on the soil antibiotic resistome[J]. Environmental Science & Technology, 2018, 52(22): 13037-13046.
(  0) 0) |
| [54] |
Liao H P, Li X, Yang Q E, et al. Herbicide selection promotes antibiotic resistance in soil microbiomes[J]. Molecular Biology and Evolution, 2021, 38(6): 2337-2350. DOI:10.1093/molbev/msab029
(  0) 0) |
| [55] |
Jia M Y, Wang F, Jin X, et al. Metal ion-oxytetracycline interactions on maize straw biochar pyrolyzed at different temperatures[J]. Chemical Engineering Journal, 2016, 304: 934-940. DOI:10.1016/j.cej.2016.05.064
(  0) 0) |
| [56] |
Cui E P, Wu Y, Jiao Y N, et al. The behavior of antibiotic resistance genes and arsenic influenced by biochar during different manure composting[J]. Environmental Science and Pollution Research International, 2017, 24(16): 14484-14490. DOI:10.1007/s11356-017-9028-z
(  0) 0) |
| [57] |
Fu Y H, Jia M Y, Wang F, et al. Strategy for mitigating antibiotic resistance by biochar and hyperaccumulators in cadmium and oxytetracycline co-contaminated soil[J]. Environmental Science & Technology, 2021, 55(24): 16369-16378.
(  0) 0) |
| [58] |
Lehmann J, Rillig M C, Thies J, et al. Biochar effects on soil biota - A review[J]. Soil Biology & Biochemistry, 2011, 43(9): 1812-1836.
(  0) 0) |
| [59] |
Gullberg E, Albrecht L M, Karlsson C, et al. Selection of a multidrug resistance plasmid by sublethal levels of antibiotics and heavy metals[J]. mBio, 2014, 5(5): e01918-e01914.
(  0) 0) |
| [60] |
Wright M S, Peltier G L, Stepanauskas R, et al. Bacterial tolerances to metals and antibiotics in metal- contaminated and reference streams[J]. FEMS Microbiology Ecology, 2006, 58(2): 293-302. DOI:10.1111/j.1574-6941.2006.00154.x
(  0) 0) |
| [61] |
Bank M S, Ok Y S, Swarzenski P W. Microplastic's role in antibiotic resistance[J]. Science, 2020, 369(6509): 1315. DOI:10.1126/science.abd9937
(  0) 0) |
| [62] |
Zhu D, Ma J, Li G, et al. Soil plastispheres as hotspots of antibiotic resistance genes and potential pathogens[J]. The ISME Journal, 2022, 16(2): 521-532. DOI:10.1038/s41396-021-01103-9
(  0) 0) |
| [63] |
Lu X M, Lu P Z, Liu X P. Fate and abundance of antibiotic resistance genes on microplastics in facility vegetable soil[J]. Science of the Total Environment, 2020, 709: 136276. DOI:10.1016/j.scitotenv.2019.136276
(  0) 0) |
| [64] |
Wu W B, Fu S S, Mao B Y, et al. Enrichment of intracellular and extracellular antibiotic resistance genes by microplastics in municipal wastewater (In Chinese)[J]. Research of Environmental Sciences, 2021, 34(6): 1434-1440. DOI:10.13198/j.issn.1001-6929.2021.02.15 [吴文斌, 付树森, 毛步云, 等. 微塑料对城市污水中胞内和胞外抗性基因的富集特征研究[J]. 环境科学研究, 2021, 34(6): 1434-1440.]
(  0) 0) |
| [65] |
Zhu D, Zheng F, Chen Q L, et al. Exposure of a soil collembolan to Ag nanoparticles and AgNO3 disturbs its associated microbiota and lowers the incidence of antibiotic resistance genes in the gut[J]. Environmental Science & Technology, 2018, 52(21): 12748-12756.
(  0) 0) |
| [66] |
Qiu Z, Yu Y, Chen Z, et al. Nanoalumina promotes the horizontal transfer of multiresistance genes mediated by plasmids across genera[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(13): 4944-4949. DOI:10.1073/pnas.1107254109
(  0) 0) |
| [67] |
McKinney C W, Pruden A. Ultraviolet disinfection of antibiotic resistant bacteria and their antibiotic resistance genes in water and wastewater[J]. Environmental Science & Technology, 2012, 46(24): 13393-13400.
(  0) 0) |
| [68] |
Qian X, Sun W, Gu J, et al. Reducing antibiotic resistance genes, integrons, and pathogens in dairy manure by continuous thermophilic composting[J]. Bioresource Technology, 2016, 220: 425-432. DOI:10.1016/j.biortech.2016.08.101
(  0) 0) |
| [69] |
Peng S, Li H J, Song D, et al. Influence of zeolite and superphosphate as additives on antibiotic resistance genes and bacterial communities during factory-scale chicken manure composting[J]. Bioresource Technology, 2018, 263: 393-401. DOI:10.1016/j.biortech.2018.04.107
(  0) 0) |
| [70] |
Cui E P, Wu Y, Zuo Y R, et al. Effect of different biochars on antibiotic resistance genes and bacterial community during chicken manure composting[J]. Bioresource Technology, 2016, 203: 11-17. DOI:10.1016/j.biortech.2015.12.030
(  0) 0) |
| [71] |
Zhang Y J, Li H C, Gu J, et al. Effects of adding different surfactants on antibiotic resistance genes and intI1 during chicken manure composting[J]. Bioresource Technology, 2016, 219: 545-551. DOI:10.1016/j.biortech.2016.06.117
(  0) 0) |
| [72] |
Zhang J Y, Wang Z Y, Wang Y W, et al. Effects of graphene oxide on the performance, microbial community dynamics and antibiotic resistance genes reduction during anaerobic digestion of swine manure[J]. Bioresource Technology, 2017, 245: 850-859. DOI:10.1016/j.biortech.2017.08.217
(  0) 0) |
| [73] |
Sun W, Gu J, Wang X J, et al. Solid-state anaerobic digestion facilitates the removal of antibiotic resistance genes and mobile genetic elements from cattle manure[J]. Bioresource Technology, 2019, 274: 287-295. DOI:10.1016/j.biortech.2018.09.013
(  0) 0) |
| [74] |
Ma Y J, Wilson C A, Novak J T, et al. Effect of various sludge digestion conditions on sulfonamide, macrolide, and tetracycline resistance genes and class I integrons[J]. Environmental Science & Technology, 2011, 45(18): 7855-7861.
(  0) 0) |
| [75] |
Zhang H C, Chang F Y, Shi P, et al. Antibiotic resistome alteration by different disinfection strategies in a full-scale drinking water treatment plant deciphered by metagenomic assembly[J]. Environmental Science & Technology, 2019, 53(4): 2141-2150.
(  0) 0) |
| [76] |
Dodd M C. Potential impacts of disinfection processes on elimination and deactivation of antibiotic resistance genes during water and wastewater treatment[J]. Journal of Environmental Monitoring, 2012, 14(7): 1754-1771. DOI:10.1039/c2em00006g
(  0) 0) |
| [77] |
Guo M T, Yuan Q B, Yang J. Distinguishing effects of ultraviolet exposure and chlorination on the horizontal transfer of antibiotic resistance genes in municipal wastewater[J]. Environmental Science & Technology, 2015, 49(9): 5771-5778.
(  0) 0) |
| [78] |
Di Cesare A, Eckert E M, D'Urso S, et al. Co-occurrence of integrase 1, antibiotic and heavy metal resistance genes in municipal wastewater treatment plants[J]. Water Research, 2016, 94: 208-214. DOI:10.1016/j.watres.2016.02.049
(  0) 0) |
| [79] |
Crofts T S, Gasparrini A J, Dantas G. Next-generation approaches to understand and combat the antibiotic resistome[J]. Nature Reviews Microbiology, 2017, 15(7): 422-434. DOI:10.1038/nrmicro.2017.28
(  0) 0) |
| [80] |
Lau C H F, van Engelen K, Gordon S, et al. Novel antibiotic resistance determinants from agricultural soil exposed to antibiotics widely used in human medicine and animal farming[J]. Applied and Environmental Microbiology, 2017, 83(16): e00989-17.
(  0) 0) |
| [81] |
Ghaly T M, Chow L, Asher A J, et al. Evolution of class 1 integrons: Mobilization and dispersal via food-borne bacteria[J]. PLoS One, 2017, 12(6): e0179169. DOI:10.1371/journal.pone.0179169
(  0) 0) |
| [82] |
Smalla K, Cook K, Djordjevic S P, et al. Environmental dimensions of antibiotic resistance: Assessment of basic science gaps[J]. FEMS Microbiology Ecology, 2018, 94(12): fiy195.
(  0) 0) |
| [83] |
Carnevali C, Morganti M, Scaltriti E, et al. Occurrence of mcr-1 in colistin-resistant Salmonella enterica isolates recovered from humans and animals in Italy, 2012 to 2015[J]. Antimicrobial Agents and Chemotherapy, 2016, 60(12): 7532-7534. DOI:10.1128/AAC.01803-16
(  0) 0) |
 2023, Vol. 60
2023, Vol. 60


