受人类活动的影响,大气中CO2浓度已较工业化前的水平增长了40%,其浓度在过去60年以每年1.7 μL·L–1快速增加[1]。大气CO2富集是全球气候变暖的主要原因之一,直接威胁到陆地生态系统的稳定性[2]。同时,气候变化会导致整个农业生产发生改变,如作物的生长环境、种类与产量等会随之变化[3]。
稻田是甲烷(CH4)的重要排放源,甲烷好氧氧化是控制稻田CH4排放的关键生物过程[4-5]。但同时水稻生长发育过程中由于淹水灌溉形成的厌氧环境,可为甲烷厌氧氧化反应的发生提供适宜条件[6]。已有研究表明,稻田土壤中存在亚硝酸盐型甲烷厌氧氧化(nitrite-dependent anaerobic methane oxidation,n-damo)反应[7-9]。该甲烷厌氧氧化反应是以NO2–作为电子受体,CH4作为其唯一电子供体[10],由与Candidatus Methylomirabilis oxyfera(M. oxyfera)相关的NC10门细菌催化完成[11]。n-damo反应及其功能微生物的发现对减缓气候变化,控制温室气体排放等方面具有重要意义,也为重新认识稻田甲烷氧化的微生物机制提供了新方向。
众多研究表明,大气CO2浓度升高影响稻田CH4排放[12-14]。CO2浓度升高可通过改变稻田土壤理化性质间接影响产甲烷菌,并导致CH4排放量增加[15]。其增加的主要原因是,大气CO2浓度升高促进水稻生长和根系分泌物的增加,从而提供了有利于产甲烷菌释放CH4的有机碳源[14,16-17]。此外,大气CO2浓度升高亦会对甲烷好氧氧化菌的群落组成和活性产生影响[13,18],继而改变稻田CH4排放。n-damo作为稻田甲烷氧化的新途径,有关其活性和功能微生物群落组成等对大气CO2浓度升高的响应知之甚少[19]。我们假设大气CO2浓度升高亦可通过影响作物生长改变土壤理化性质,从而间接对n-damo过程产生影响。值得注意的是,大气CO2浓度的增加并不是瞬时增加至某一浓度,而是随时间推移缓慢增加的过程。但目前,少有研究报道CO2浓度缓增对稻田土壤甲烷氧化菌的影响[18]。
为更好地研究气候变化对n-damo过程的影响,更真实地模拟大气CO2的变化,本研究设置CO2缓增处理(每年增加40 μL·L–1,直至增幅达160 μL·L–1),从M. oxyfera-like细菌的群落组成、多样性、丰度和n-damo活性等多方面分析大气CO2浓度升高是如何作用于稻田n-damo过程。研究结果对预测未来CO2浓度升高对n-damo的影响具有重要意义。
1 材料与方法 1.1 试验区概况试验区位于江苏省南京市农业气象与生态试验站的稻田试验地(32.16°N,118.86°E),亚热带湿润气候,年平均降水量1 110 mm,年平均气温15.6℃。供试土壤为潴育型水稻土,耕层土壤为壤质黏土。稻田氮肥年施用量为N 250 kg·hm–2,水分管理方式为在水稻生育前、中、后期分别进行淹水、烤田、干湿交替处理[15]。
1.2 试验设计研究依托2016年建成并运行至今的开顶式气室(Open top chambers,OTC)组成的CO2浓度升高自动调控平台,模拟大气CO2浓度[20]。每个OTC配备CO2传感器(GMM222传感器,芬兰)和温湿度自动记录仪。OTC中CO2浓度通过计算机程序实现自动监测和调整,以确保CO2浓度保持在目标浓度值。以大气CO2浓度作为对照组(CK),以CO2浓度缓增作为处理组(EC),每个处理均设置3个重复。EC处理组为在CK基础上,自2016年起每年增加40 μL·L–1,至采样时增幅达160 μL·L–1。
2019年,分别在水稻分蘖期、拔节期、扬花期采集0~20 cm土壤样品。使用直径5 cm的圆柱形取样器,利用五点采样法采集每个OTC内的土柱,按0~5 cm、5~10 cm、10~20 cm分段,并对同一OTC内同一深度样品均匀混合,储存在无菌密封袋中,冷藏运送至实验室。
1.3 土壤理化因子分析土壤理化性质参照《土壤农化分析》[21]中的方法测定。采用烘干法测定土壤含水量(Water content,WC)。采用pH计法测定土壤pH,土水比1﹕2.5(m/v)。采用重铬酸钾—外加热法测定土壤有机碳(Soil organic carbon,SOC)含量。采用比色法测定经蒸馏水浸提后浸提液中可溶性有机碳(Dissolved organic carbon,DOC)浓度。采用分光光度法测定经2 mol·L–1 KCl浸提后浸提液中NO2–、NO3–、NH4+浓度。
1.4 亚硝酸盐型甲烷厌氧氧化活性测定采用厌氧泥浆培养法测定亚硝酸盐型甲烷厌氧氧化活性[19]。将一定量土壤样品与去离子水转移至玻璃瓶中制成泥浆,并对玻璃瓶进行厌氧处理。随后对泥浆进行为期3 d的预培养。预培养结束后,设置两组同位素示踪:(1)13CH4,(2)13CH4+ NO2–。用13CH4(13C丰度99.9%)进行等体积气体置换,使得瓶内13CH4体积占比约10%。此外,13CH4+ NO2–处理加入0.05 mL的NO2–浓缩液,使瓶中NO2–的初始浓度维持在0.5 mmol·L–1;13CH4处理加入0.05 mL的蒸馏水。随后继续将玻璃瓶置于摇床培养。分别在第0、7、14和21天向对应瓶子中加入50%的ZnCl2溶液以终止反应。采用同位素比质谱仪(Isoprime 100,英国)联合气相色谱仪(Agilent 7890B,美国),测定顶空气体中的13CO2。根据玻璃瓶中13CO2产生量随时间的改变计算n-damo活性。
1.5 DNA提取,PCR扩增与高通量测序使用E.Z.N.A.® soil DNA kit(Omega Bio-tek,美国)提取不同CO2处理下水稻土壤样品的总DNA。用超微量分光光度计(NanoDrop2000)检测DNA的浓度和纯度,用1%琼脂糖凝胶电泳检测DNA的质量。
对M. oxyfera-like细菌的16S rRNA基因进行巢式PCR扩增(PCR仪器为ABI GeneAmp®9700)。第一步扩增选用引物202F/1545R;第二步扩增选用引物qp1f/qp2r[22]。PCR反应体系为20 µL,包含4 µL 5×FastPfu缓冲液、2 µL 2.5 mm dNTPs、0.8 µL引物(5 µmol·L–1)、0.4 µL FastPfu聚合酶和10 ng DNA模板。PCR热反应条件如下:95℃预变性3 min;95℃变性1 min,63℃退火1 min,72℃延伸1 min,30个循环;最后72℃延伸10 min。利用AxyPrep DNA Gel Extraction Kit(Axygen Biosciences,Union City,CA,美国)对PCR产物进行纯化。通过Illumina MiSeq平台(Illumina,San Diego,美国)将纯化后的扩增片段构建PE300文库。
1.6 序列分析利用FLASH软件对原始序列进行筛选和拼接。使用UPARSE软件,以97%的相似性对序列进行OTU(Operational taxonomic unit)聚类,同时在聚类过程中去除单个序列和嵌合体。利用RDP Classifier对所获得的序列进行注释。此外,通过将OTU代表性序列与NCBI数据库进行比较,进一步去除非M. oxyfera-like细菌序列,以确保序列分析的准确性。采用Shannon指数和Chao1指数对M. oxyfera-like细菌多样性进行分析。
1.7 系统发育分析使用BLAST在NCBI数据库中搜索M. oxyfera- like细菌优势OTU的相似序列,随后用MEGA-X软件的邻接方法构建包含优势OTU代表性序列及其相似序列的系统发育树。
1.8 定量PCR使用特异性引物qP1F/qP1R[22]对M. oxyfera- like细菌的16S rRNA基因进行定量PCR。PCR热反应条件如下:95℃条件下预变性3 min;95℃下变性0.5 min,63℃下退火0.5 min,72℃下延伸0.5 min,40个循环;最后72℃下延伸5 min。将已知含有16S rRNA基因的质粒DNA稀释10倍,构建标准曲线(每个浓度重复3次)。根据标准曲线计算每个样品中M. oxyfera-like细菌的丰度。
1.9 统计分析分别采用主坐标分析(Principal co-ordinates analysis,PCoA)和冗余分析(Redundancy analysis,RDA)检验不同处理间M. oxyfera-like细菌群落的差异及其与环境因子间的相互作用关系。采用SPSS 25.0软件进行t检验,分析不同CO2处理间n-damo活性,以及M. oxyfera-like细菌丰度、多样性等各项指标的差异及显著性。利用Pearson相关性分析检验各理化指标与微生物指标间的关系。利用OriginPro 2021、R 4.0.5等软件绘图。
2 结果 2.1 不同CO2处理下土壤理化性质稻田土壤样品的理化性质如表 1所示。由表可知,除拔节期10~20 cm土层外,不同CO2处理下SOC变化不显著。土壤pH在不同CO2处理下无显著变化。土壤含水量随土层深度的增加呈降低趋势,且EC处理显著降低了分蘖期5~10 cm、拔节期10~20 cm、扬花期10~20 cm土壤的含水量(P < 0.05)。EC处理下分蘖期各土层中NO2–-N含量显著低于CK(P < 0.05),但其余时期未有显著变化。土壤NO3–-N含量在分蘖期0~5 cm、拔节期0~5 cm、扬花期0~5 cm、10~20 cm土层无显著变化,其余时期CK处理均显著高于EC(P < 0.05)。伴随水稻的生长,土壤NH4+-N含量呈下降趋势,且仅在分蘖期10~20 cm、拔节期5~20 cm、扬花期0~5 cm土层无显著变化,而其余时期EC处理均显著低于CK(P < 0.05)。EC处理显著增加了扬花期5~10 cm土层中的DOC含量(P < 0.05)。
|
|
表 1 不同CO2浓度处理下各生育期稻田土壤理化因子 Table 1 Physiochemical properties of paddy soils during different rice growth stages under different CO2 treatments |
如图 1A所示,在分蘖期,EC处理0~5 cm土层的n-damo活性显著增加(P < 0.05),但5~10 cm的活性显著降低(P < 0.05)。EC处理分蘖期n-damo活性整体降低了8%(P > 0.05)。EC处理显著增加了拔节期0~10 cm土层的n-damo活性(P < 0.05),但显著降低了10~20 cm的活性(P < 0.05)。EC处理使拔节期n-damo活性整体增幅达137.9%(P < 0.05)。在扬花期,EC处理虽使0~5 cm、10~20 cm土层n-damo活性显著增加(P < 0.05),却使5~10 cm活性显著降低(P < 0.05)。扬花期EC处理下n-damo活性较CK整体增加了23.0%(P > 0.05)。
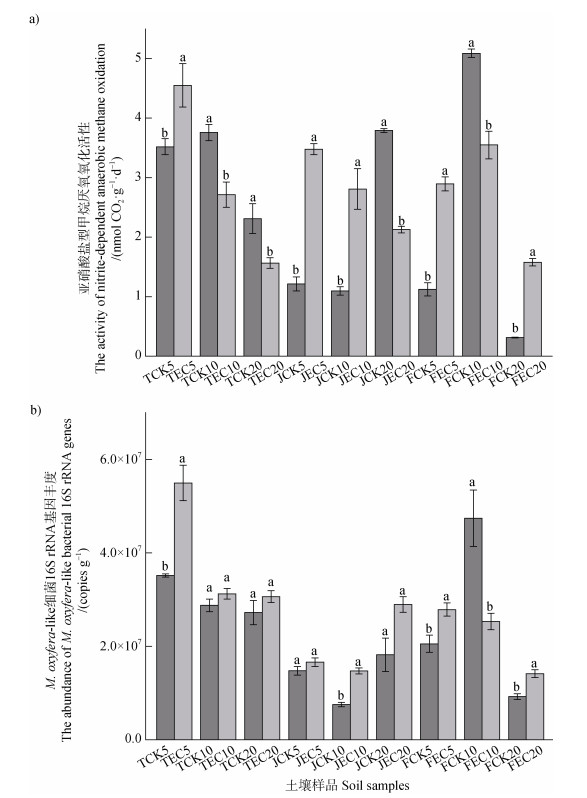
|
注:1)TCK,分蘖期CK处理;TEC,分蘖期EC处理;JCK,拔节期CK处理;JEC,拔节期EC处理;FCK,扬花期CK处理;FEC,扬花期EC处理;其后数字5、10、20分别表示0~5 cm、5~10 cm、10~20 cm土层。2)图中误差线为标准误;不同字母表示该生育期下同一深度不同CO2处理间存在显著性差异(P < 0.05)。 Note: 1)TCK, CK treatment at the tillering stage; TEC, EC treatment at the tillering stage; JCK, CK treatment at the jointing stage; JEC, EC treatment at the jointing stage; FCK, CK treatment at the flowering stage; FEC, EC treatment at the flowering stage; the numbers of 5, 10, and 20 represent soil layers of 0~5 cm, 5~10 cm, and 10~20 cm, respectively. 2)Error bars in the figure are standard errors. Different letters indicate a significant difference between CO2 treatments at the same growth stage and depth (P < 0.05). 图 1 不同CO2浓度处理下各生育期不同深度稻田土壤n-damo活性与M. oxyfera-like细菌丰度 Fig. 1 The n-damo activity and abundance of M. oxyfera-like bacteria in different depths of paddy soil across different growth stages under different CO2 treatments |
如图 1B所示,EC处理显著增加了分蘖期0~5 cm土层中M. oxyfera-like细菌的16S rRNA基因丰度(P < 0.05),但显著降低了扬花期5~10 cm的基因丰度(P < 0.05)。相较于CK,EC处理使分蘖期基因丰度整体增加了28%(P > 0.05)。EC处理显著增加了拔节期5~10 cm土层中的基因丰度(P < 0.05),同时使该时期基因丰度的整体增幅达96.0%(P < 0.05)。在扬花期,EC处理使0~5 cm、10~20 cm土层中基因丰度显著增加(P < 0.05),但使5~10 cm的基因丰度显著降低(P < 0.05)。EC处理使扬花期基因丰度整体降低13%(P > 0.05)。
2.3 不同CO2处理下土壤中M. oxyfera-like细菌多样性M. oxyfera-like细菌的16S rRNA基因多样性如表 2所示。由表可知,单个样品的覆盖度均超过99%,说明本研究采用的高通量测序方法能较好地表征此类微生物的基因多样性。EC处理下的OTU数、Shannon指数和Chao1指数分别为6~12、0.79~1.45、6.0~12.0;而CK处理组的数据分别为10~17、0.70~1.61、10.0~17.0。相较于CK处理,EC显著提高了OTU数(P < 0.05)。
|
|
表 2 不同CO2处理下各生育期不同深度稻田土壤中M. oxyfera-like细菌16S rRNA基因的多样性 Table 2 Diversity of M. oxyfera-like bacterial 16S rRNA genes in different depths of paddy soils across different growth stages under different CO2 treatments |
M. oxyfera-like细菌的优势OTU(占总序列的94.1%)落入两个不同的基因簇(图 2)。Cluster A和Cluster B中的序列与已知的M. oxyfera的16S rRNA基因相似度分别为92.8%~96.4%和92.8%~94.2%。此外,对M. oxyfera-like细菌的群落结构进行PCoA分析发现,不同CO2浓度处理下其群落组成存在显著差异(P < 0.05)(图 3a)。组间差异分析结果显示,EC处理下OTU51和OTU26的相对丰度较CK处理有显著变化(P < 0.05)(图 3b)。
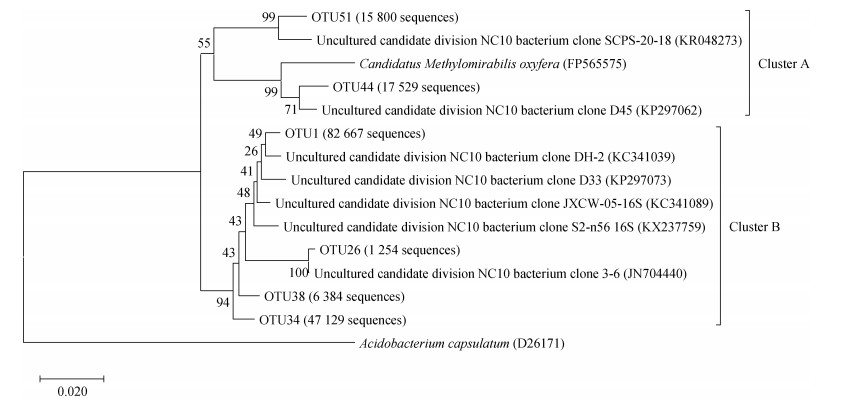
|
图 2 稻田土壤中优势OTUs序列的系统发育树 Fig. 2 Phylogenetic trees showing the affiliation of the dominant OTUs obtained from the paddy soil |
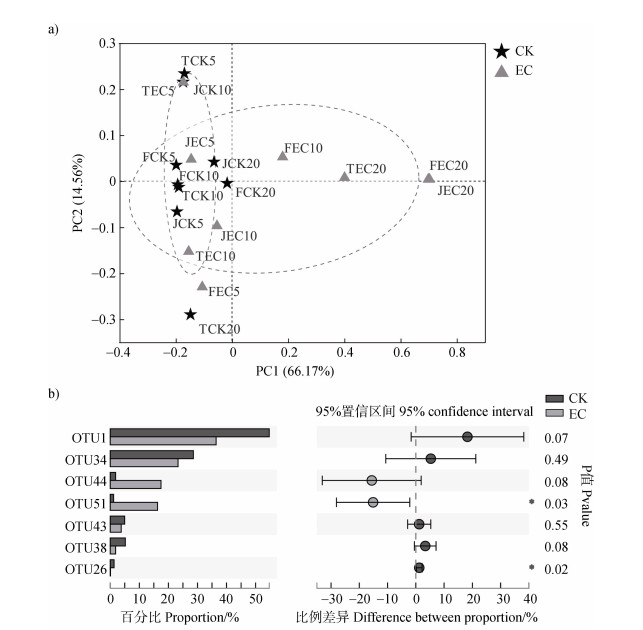
|
图 3 不同CO2处理下M. oxyfera-like细菌群落组成的主坐标分析(a)及其组间差异(b) Fig. 3 Principal co-ordinates analysis of M. oxyfera-like bacterial communities under different CO2 treatments(a)and the difference in community composition between treatments(b) |
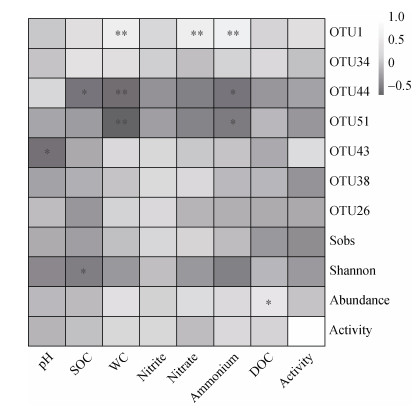
冗余分析结果显示,土壤WC、NO3–和NH4+含量对稻田土壤中M. oxyfera-like细菌的群落结构均有显著影响(P < 0.05)(图 4)。相关性分析表明,土壤DOC含量与M. oxyfera-like细菌丰度呈显著正相关(P < 0.05),而SOC含量与Shannon指数呈显著负相关(P < 0.05)(图 5)。土壤pH与OTU41的相对丰度呈显著负相关(P < 0.05)。此外,OTU1与WC、NO3–和NH4+含量呈极显著正相关(P < 0.01)。OTU44与NH4+和SOC含量呈显著负相关(P < 0.05);OTU51与NH4+含量呈显著负相关(P < 0.05)。此外,OTU51与OTU44均与WC呈极显著负相关(P < 0.01)。
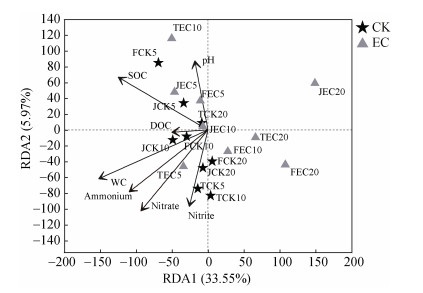
|
图 4 不同CO2处理下M. oxyfera-like细菌群落组成与环境因子的冗余分析 Fig. 4 Redundancy analysis of M. oxyfera-like bacteria community composition and environmental factors under different CO2 treatments |
|
注:白色为正相关,灰色为负相关,颜色深浅代表相关系数大小;“*”表示具有显著相关性(P < 0.05),“* *”表示具有极显著相关性(P < 0.01)。 Note: The white represents positive correlation while the grey represents negative correlation. The color depth denotes the size of the correlation coefficient. "*" means significant correlation(P < 0.05)and "* *" means extremely significant correlation(P < 0.01). 图 5 n-damo活性及M. oxyfera-like细菌16S rRNA基因丰度、各优势OTUs丰度、多样性与土壤理化性质间的相关性热图 Fig. 5 The heatmap showing correlations among the n-damo activity, 16S rRNA gene abundance of M. oxyfera-like bacteria, the relative abundance of dominant OTUs, diversity and the soil physicochemical properties |
同位素示踪结果显示,不同CO2处理下各生育期不同深度稻田土壤中均存在n-damo活性。本研究中n-damo活性为0.31~5.09 nmol·g–1·d–1(以CO2计,下同),在先前稻田(0.2~11.5 nmol·g–1·d–1)[8,23]、天然湿地(0.2~14.5 nmol·g–1·d–1)[24-26]中报道的n-damo活性范围之内,但低于部分人工湿地(8.5~23.5 nmol·g–1·d–1)和内陆河流(0.4~61.0 nmol·g–1·d–1)[27-28]的报道值。本研究中M. oxyfera-like细菌的16S rRNA基因丰度范围为7.51×106~5.49×107 copies·g–1。这些值高于中国亚热带地区稻田土壤(6.5×103~9.3×106 copies·g–1)[29-30],以及部分天然湿地(104~ 105 copies·g–1)[31]、城市湿地(1.6×106~1.3×107 copies·g–1)[32]中报道的丰度值。此外,所试土壤中n-damo活性与M. oxyfera-like细菌的基因丰度无显著正相关(图 5)。其原因可能是本文针对的目标基因为M. oxyfera-like细菌的16S rRNA基因,并非其功能基因。因此,未来有必要针对稻田土壤中M. oxyfera-like细菌的功能基因(如pmoA[22])丰度与n-damo活性间的关系展开相关研究。
本研究发现,大气CO2升高(缓增)处理对稻田土壤n-damo活性及其功能微生物M. oxyfera-like细菌丰度均有重要影响。大气CO2升高条件下,稻田n-damo活性整体提高13.7%,M. oxyfera-like细菌丰度整体增加17.0%。虽两者的整体增幅并未达到显著水平,但EC处理使拔节期n-damo活性与M. oxyfera-like细菌丰度均有显著提高(P < 0.05)。相关性分析表明,M. oxyfera-like细菌丰度与土壤DOC含量呈显著正相关(P < 0.05)(图 5)。大气CO2浓度升高可通过影响水稻生长和根系分泌物的增加,间接导致土壤中DOC含量的增加[33]。DOC含量通常与CH4含量呈正相关关系[15],其原因是DOC含量增加使产甲烷菌有更多的底物来源,促进其CH4的产生[17]。土壤中CH4含量的增加会为M. oxyfera-like细菌提供更多的电子供体,进而可能促进其生长与代谢。Shen等[28]研究发现,缺氧河床沉积物中n-damo活性和M. oxyfera-like细菌丰度均与CH4含量呈显著正相关。此外,DOC含量的增加还会增强土壤反硝化活性[34],因而会加速土壤中NO3–的还原,从而为M. oxyfera-like细菌提供更多的NO2–来源。M. oxyfera-like细菌正是利用NO2–为电子受体对CH4进行氧化,并将NO2–还原为N2[11,35]。本研究发现,大气CO2浓度升高使得所试稻田土壤中NO3–含量在大部分生育期和深度下显著降低(表 1),其重要原因可能是CO2浓度升高促进了反硝化活性[36]。此外,EC处理使稻田土壤NH4+含量明显降低(表 1),其原因之一可能是大气CO2浓度的增加使得土壤硝化活性增强[37]。硝化活性的增强亦可为M. oxyfera-like细菌提供更多的NO2–来源。综上,大气CO2浓度升高通过影响作物生长导致土壤DOC含量增加,并通过改变相关微生物过程对n-damo活性及M. oxyfera-like细菌丰度产生间接影响。
通过高通量测序手段发现,不同处理下各生育期不同深度稻田土壤中均存在M. oxyfera-like细菌序列。所获序列与已知M. oxyfera的16S rRNA基因相似度为92.8%~96.4%。系统发育分析表明(图 2),Cluster A中的序列与黄河口泥沙[38]、水稻土(NCBI登录号为KR048273,unpublished)中报道的NC10门序列具有较高亲缘性(相似度分别为92.8%~98.8%和94.2%~98.6%)。Cluster B包含的序列占比超过75%,该基因簇中的序列与东湖沉积物[31]、黄河口泥沙[38]、水稻土[30]、泥炭地(NCBI登录号为KX237759,unpublished)中报道的NC10门序列高度相似(相似度分别为97.4%~99.0%、96.9%~100%、97.4%~98.6%、97.1%~98.8%)。多样性分析结果显示,单个土壤样品中M. oxyfera- like细菌16S rRNA基因的OTUs数为6~17,高于先前稻田土壤中的报道值(3~6 OTUs)[23,39]。
本研究还发现,大气CO2浓度升高对稻田土壤中M. oxyfera-like细菌的群落组成具有显著影响(P < 0.05)(图 3A)。大气CO2浓度升高使得本研究中各生育期部分深度土壤中WC、NO3–和NH4+含量发生了显著变化(P < 0.05)(表 1)。前人研究也表明,CO2浓度升高会导致土壤中WC和无机氮水平发生改变[15,38]。Tian等[19]与Hui等[39]研究发现,WC和NO3–含量对稻田土壤中M. oxyfera-like细菌的群落组成和多样性有显著影响。Chen等[40]和Shen等[35]研究表明,NH4+和NO3–含量是影响湿地沉积物中M. oxyfera-like细菌群落结构的重要环境因子。由于M. oxyfera-like细菌是一类厌氧菌,其对于氧气浓度的变化较为敏感[41]。Wang等[42]研究发现,累积淹水时间对淡水沉积物中M. oxyfera-like细菌群落结构有重要影响。因此,WC很可能通过影响稻田土壤中氧气浓度对M. oxyfera-like细菌群落结构产生影响。此外,土壤中NH4+和NO3–含量的变化会改变硝化和反硝化作用强度,继而通过影响NO2–的供给对M. oxyfera-like细菌产生影响。
4 结论大气CO2浓度升高在一定程度上提高了稻田n-damo活性和M. oxyfera-like细菌丰度,且显著改变了此类细菌的群落结构。大气CO2浓度升高增加了土壤中可溶性有机碳含量,并通过改变CH4和NO2–的供给,进一步影响M. oxyfera-like细菌的生长和代谢。此外,土壤中含水量、NO3–和NH4+含量的变化对M. oxyfera-like细菌群落组成有显著影响。
| [1] |
Jin J, Wood J, Franks A, et al. Long-term CO2 enrichment alters the diversity and function of the microbial community in soils with high organic carbon[J]. Soil Biology & Biochemistry, 2020, 144: 107780.
(  0) 0) |
| [2] |
Liu S W, Ji C, Wang C, et al. Climatic role of terrestrial ecosystem under elevated CO2: A bottom-up greenhouse gases budget[J]. Ecology Letters, 2018, 21(7): 1108-1118. DOI:10.1111/ele.13078
(  0) 0) |
| [3] |
van Meijl H, Havlik P, Lotze-Campen H, et al. Comparing impacts of climate change and mitigation on global agriculture by 2050[J]. Environmental Research Letters, 2018, 13(6): 064021. DOI:10.1088/1748-9326/aabdc4
(  0) 0) |
| [4] |
le Mer J, Roger P. Production, oxidation, emission and consumption of methane by soils: A review[J]. European Journal of Soil Biology, 2001, 37(1): 25-50. DOI:10.1016/S1164-5563(01)01067-6
(  0) 0) |
| [5] |
Kirschke S, Bousquet P, Ciais P, et al. Three decades of global methane sources and sinks[J]. Nature Geoscience, 2013, 6(10): 813-823. DOI:10.1038/ngeo1955
(  0) 0) |
| [6] |
Shen L D. A review of study on microbial ecology of nitrite-dependent anaerobic methane oxidation (In Chinese)[J]. Acta Pedologica Sinica, 2015, 52(4): 713-722. [沈李东. 亚硝酸盐型甲烷厌氧氧化微生物生态学研究进展[J]. 土壤学报, 2015, 52(4): 713-722.]
(  0) 0) |
| [7] |
Vaksmaa A, Lüke C, van Alen T, et al. Distribution and activity of the anaerobic methanotrophic community in a nitrogen-fertilized Italian paddy soil[J]. FEMS Microbiology Ecology, 2016, 92(12): fiw181. DOI:10.1093/femsec/fiw181
(  0) 0) |
| [8] |
Shen L D, Liu J Q, Yang Y L, et al. Activity, abundance and community composition of nitrite-dependent methanotrophs in response to fertilization in paddy soils[J]. Applied Soil Ecology, 2021, 166: 103987. DOI:10.1016/j.apsoil.2021.103987
(  0) 0) |
| [9] |
Fan L C, Shahbaz M, Ge T D, et al. To shake or not to shake: 13C-based evidence on anaerobic methane oxidation in paddy soil[J]. Soil Biology & Biochemistry, 2019, 133: 146-154.
(  0) 0) |
| [10] |
Raghoebarsing A A, Pol A, van de Pas-Schoonen K T, et al. A microbial consortium couples anaerobic methane oxidation to denitrification[J]. Nature, 2006, 440(7086): 918-921. DOI:10.1038/nature04617
(  0) 0) |
| [11] |
Ettwig K F, Butler M K, le Paslier D, et al. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria[J]. Nature, 2010, 464(7288): 543-548. DOI:10.1038/nature08883
(  0) 0) |
| [12] |
Xu Z J, Zheng X H, Wang Y S, et al. Effects of elevated CO2 and N fertilization on CH4 emissions from paddy rice fields[J]. Global Biogeochemical Cycles, 2004, 18(3): GB3009.
(  0) 0) |
| [13] |
Liu Y, Liu X Y, Cheng K, et al. Responses of methanogenic and methanotrophic communities to elevated atmospheric CO2 and temperature in a paddy field[J]. Frontiers in Microbiology, 2016, 7: 1895.
(  0) 0) |
| [14] |
Tokida T, Fumoto T, Cheng W, et al. Effects of free-air CO2 enrichment(FACE)and soil warming on CH4 emission from a rice paddy field: Impact assessment and stoichiometric evaluation[J]. Biogeosciences, 2010, 7(9): 2639-2653. DOI:10.5194/bg-7-2639-2010
(  0) 0) |
| [15] |
Wang Y Y, Hu Z H, Shen L D, et al. The process of methanogenesis in paddy fields under different elevated CO2 concentrations[J]. Science of the Total Environment, 2021, 773: 145629. DOI:10.1016/j.scitotenv.2021.145629
(  0) 0) |
| [16] |
Yang L X, Wang Y L, Huang J Y, et al. Seasonal changes in the effects of free-air CO2 enrichment(FACE)on phosphorus uptake and utilization of rice at three levels of nitrogen fertilization[J]. Field Crops Research, 2007, 102(2): 141-150. DOI:10.1016/j.fcr.2007.03.004
(  0) 0) |
| [17] |
Okubo T, Liu D Y, Tsurumaru H, et al. Elevated atmospheric CO2 levels affect community structure of rice root-associated bacteria[J]. Frontiers in Microbiology, 2015, 6: 136.
(  0) 0) |
| [18] |
Liu X, Shen L D, Tian M H, et al. Effect of slow increase of atmospheric CO2 concentration on methane oxidation in paddy soils (In Chinese)[J]. Acta Pedologica Sinica, 2022, 59(2): 568-579. [刘心, 沈李东, 田茂辉, 等. 大气CO2浓度缓增对稻田土壤甲烷氧化过程的影响[J]. 土壤学报, 2022, 59(2): 568-579.]
(  0) 0) |
| [19] |
Tian M H, Shen L D, Liu X, et al. Response of nitrite-dependent anaerobic methanotrophs to elevated atmospheric CO2 concentration in paddy fields[J]. Science of the Total Environment, 2021, 801: 149785. DOI:10.1016/j.scitotenv.2021.149785
(  0) 0) |
| [20] |
Liu C, Hu Z H, Yu L F, et al. Responses of photosynthetic characteristics and growth in rice and winter wheat to different elevated CO2 concentrations[J]. Photosynthetica, 2020, 58(5): 1130-1140. DOI:10.32615/ps.2020.066
(  0) 0) |
| [21] |
Bao S D. Soil and agricultural chemistry analysis (In Chinese). Beijing: China Agriculture Press, 2000. [鲍士旦. 土壤农化分析[M]. 北京: 中国农业出版社, 2000.]
(  0) 0) |
| [22] |
Ettwig K F, van Alen T, van de Pas-Schoonen K T, et al. Enrichment and molecular detection of denitrifying methanotrophic bacteria of the NC10 Phylum[J]. Applied and Environmental Microbiology, 2009, 75(11): 3656-3662. DOI:10.1128/AEM.00067-09
(  0) 0) |
| [23] |
Shen L D, Liu S, Huang Q, et al. Evidence for the cooccurrence of nitrite-dependent anaerobic ammonium and methane oxidation processes in a flooded paddy field[J]. Applied and Environmental Microbiology, 2014, 80(24): 7611-7619. DOI:10.1128/AEM.02379-14
(  0) 0) |
| [24] |
Shen L D, Huang Q, He Z F, et al. Vertical distribution of nitrite-dependent anaerobic methane-oxidising bacteria in natural freshwater wetland soils[J]. Applied Microbiology and Biotechnology, 2015, 99(1): 349-357. DOI:10.1007/s00253-014-6031-x
(  0) 0) |
| [25] |
Hu B L, Shen L D, Lian X, et al. Evidence for nitrite- dependent anaerobic methane oxidation as a previously overlooked microbial methane sink in wetlands[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(12): 4495-4500.
(  0) 0) |
| [26] |
Xie F, Ma A Z, Zhou H C, et al. Niche differentiation of denitrifying anaerobic methane oxidizing bacteria and Archaea leads to effective methane filtration in a Tibetan alpine wetland[J]. Environment International, 2020, 140: 105764. DOI:10.1016/j.envint.2020.105764
(  0) 0) |
| [27] |
Zhang M P, Huang J C, Sun S S, et al. Depth-specific distribution and significance of nitrite-dependent anaerobic methane oxidation process in tidal flow constructed wetlands used for treating river water[J]. Science of the Total Environment, 2020, 716: 137054. DOI:10.1016/j.scitotenv.2020.137054
(  0) 0) |
| [28] |
Shen L D, Ouyang L, Zhu Y Z, et al. Active pathways of anaerobic methane oxidation across contrasting riverbeds[J]. The ISME Journal, 2019, 13(3): 752-766. DOI:10.1038/s41396-018-0302-y
(  0) 0) |
| [29] |
Shen L D, Wu H S, Gao Z Q, et al. Presence of diverse Candidatus Methylomirabilis oxyfera-like bacteria of NC10 Phylum in agricultural soils[J]. Journal of Applied Microbiology, 2016, 120(6): 1552-1560. DOI:10.1111/jam.13119
(  0) 0) |
| [30] |
Wang Y, Zhu G B, Harhangi H R, et al. Co-occurrence and distribution of nitrite-dependent anaerobic ammonium and methane-oxidizing bacteria in a paddy soil[J]. FEMS Microbiology Letters, 2012, 336(2): 79-88. DOI:10.1111/j.1574-6968.2012.02654.x
(  0) 0) |
| [31] |
Zhu G B, Zhou L L, Wang Y, et al. Biogeographical distribution of denitrifying anaerobic methane oxidizing bacteria in Chinese wetland ecosystems[J]. Environmental Microbiology Reports, 2015, 7(1): 128-138. DOI:10.1111/1758-2229.12214
(  0) 0) |
| [32] |
Segarra K E A, Schubotz F, Samarkin V, et al. High rates of anaerobic methane oxidation in freshwater wetlands reduce potential atmospheric methane emissions[J]. Nature Communications, 2015, 6: 7477. DOI:10.1038/ncomms8477
(  0) 0) |
| [33] |
Sun X, Han X G, Ping F, et al. Effect of rice-straw biochar on nitrous oxide emissions from paddy soils under elevated CO2 and temperature[J]. Science of the Total Environment, 2018, 628/629: 1009-1016. DOI:10.1016/j.scitotenv.2018.02.046
(  0) 0) |
| [34] |
Qin S P, Hu C S, Clough T J, et al. Irrigation of DOC-rich liquid promotes potential denitrification rate and decreases N2O/(N2O+N2)product ratio in a 0-2 m soil profile[J]. Soil Biology & Biochemistry, 2017, 106: 1-8.
(  0) 0) |
| [35] |
Shen L D, Wu H S, Liu X, et al. Cooccurrence and potential role of nitrite- and nitrate-dependent methanotrophs in freshwater marsh sediments[J]. Water Research, 2017, 123: 162-172. DOI:10.1016/j.watres.2017.06.075
(  0) 0) |
| [36] |
Carnol M, Hogenboom L, Jach M E, et al. Elevated atmospheric CO2 in open top Chambers increases net nitrification and potential denitrification[J]. Global Change Biology, 2002, 8(6): 590-598. DOI:10.1046/j.1365-2486.2002.00493.x
(  0) 0) |
| [37] |
Shen L D, Yang Y L, Liu J Q, et al. Different responses of ammonia-oxidizing Archaea and bacteria in paddy soils to elevated CO2 concentration[J]. Environmental Pollution, 2021, 286: 117558. DOI:10.1016/j.envpol.2021.117558
(  0) 0) |
| [38] |
Yan P Z, Li M C, Wei G S, et al. Molecular fingerprint and dominant environmental factors of nitrite-dependent anaerobic methane-oxidizing bacteria in sediments from the Yellow River Estuary, China[J]. PLoS One, 2015, 10(9): e0137996. DOI:10.1371/journal.pone.0137996
(  0) 0) |
| [39] |
Hui C, Guo X X, Sun P F, et al. Depth-specific distribution and diversity of nitrite-dependent anaerobic ammonium and methane-oxidizing bacteria in upland- cropping soil under different fertilizer treatments[J]. Applied Soil Ecology, 2017, 113: 117-126. DOI:10.1016/j.apsoil.2017.02.005
(  0) 0) |
| [40] |
Chen J, Zhou Z C, Gu J D. Complex community of nitrite-dependent anaerobic methane oxidation bacteria in coastal sediments of the Mai Po wetland by PCR amplification of both 16S rRNA and pmoA genes[J]. Applied Microbiology and Biotechnology, 2015, 99(3): 1463-1473. DOI:10.1007/s00253-014-6051-6
(  0) 0) |
| [41] |
Luesken F A, Wu M L, den Camp H J M O, et al. Effect of oxygen on the anaerobic methanotroph 'Candidatus Methylomirabilis oxyfera': Kinetic and transcriptional analysis[J]. Environmental Microbiology, 2012, 14(4): 1024-1034. DOI:10.1111/j.1462-2920.2011.02682.x
(  0) 0) |
| [42] |
Wang Y, Huang P, Ye F, et al. Nitrite-dependent anaerobic methane oxidizing bacteria along the water level fluctuation zone of the Three Gorges Reservoir[J]. Applied Microbiology and Biotechnology, 2016, 100(4): 1977-1986. DOI:10.1007/s00253-015-7083-2
(  0) 0) |
 2023, Vol. 60
2023, Vol. 60


